Abstract
BACKGROUND AND PURPOSE: Total brain volume and total intracranial volume are important measures for assessing whole-brain atrophy in Alzheimer disease, dementia, and other neurodegenerative diseases. Unlike MR imaging, which has a number of well-validated fully-automated methods, only a handful of methods segment CT images. Available methods either use enhanced CT, do not estimate both volumes, or require formal validation. Reliable computation of total brain volume and total intracranial volume from CT is needed because head CTs are more widely used than head MRIs in the clinical setting. We present an automated head CT segmentation method (CTseg) to estimate total brain volume and total intracranial volume.
MATERIALS AND METHODS: CTseg adapts a widely used brain MR imaging segmentation method from the Statistical Parametric Mapping toolbox using a CT-based template for initial registration. CTseg was tested and validated using head CT images from a clinical archive.
RESULTS: CTseg showed excellent agreement with 20 manually segmented head CTs. The intraclass correlation was 0.97 (P < .001) for total intracranial volume and 0.94 (P < .001) for total brain volume. When CTseg was applied to a cross-sectional Alzheimer disease dataset (58 with Alzheimer disease patients and 58 matched controls), CTseg detected a loss in percentage total brain volume (as a percentage of total intracranial volume) with age (P < .001) as well as a group difference between patients with Alzheimer disease and controls (P < .01). We observed similar results when total brain volume was modeled with total intracranial volume as a confounding variable.
CONCLUSIONS: In current clinical practice, brain atrophy is assessed by inaccurate and subjective “eyeballing” of CT images. Manual segmentation of head CT images is prohibitively arduous and time-consuming. CTseg can potentially help clinicians to automatically measure total brain volume and detect and track atrophy in neurodegenerative diseases. In addition, CTseg can be applied to large clinical archives for a variety of research studies.
ABBREVIATIONS:
- AD
- Alzheimer disease
- BET
- Brain Extraction Tool
- ICC
- intraclass correlation coefficient
- TBV
- total brain volume
- TIV
- total intracranial volume
- TPM
- tissue probability map
Total brain volume (TBV) is an important measure for assessing brain atrophy in aging and neurodegenerative diseases.1 TBV is estimated from MR or x-ray CT brain images by segmenting the brain parenchyma using manual or automated methods. Automated methods are preferred due to efficiency, reliability, and reproducibility. A number of automated segmentation methods are available for MR images that are extensively applied in the clinical domain.2 In the clinical setting, CT is more widely used than MR imaging due to faster acquisition speed, a smaller number of contraindications, lower cost, and its ability to answer a range of clinical questions.3 However, only a handful of automated segmentation methods exist for head CT images.
Several existing methods in CT segmentation are either semiautomated4,5 or targeted toward a specific brain region6⇓-8 or disease condition.5,9 Methods available for segmenting CT images to measure global volume metrics such as total intracrancial volume (TIV) and TBV from images with no detectable pathology were not formally validated.10⇓-12 Some well-validated methods segment only TIV13,14 but not TBV. However, TBV is more indicative of disease conditions in neurodegenerative dieseases,15 and TIV is used merely as a nuisance variable for normalization purposes. Manniesing et al4 estimated TBV using head CT but used enhanced CT images.4 However, their method cannot be applied to single-time-point CT images with no image enhancement. Irimia et al16 adapted SPM12 (http://www.fil.ion.ucl.ac.uk/spm/),17 a widely used MR imaging–based segmentation method, for CT segmentation and validated it by comparing it with MR images segmented using FreeSurfer (http://surfer.nmr.mgh.harvard.edu). However, they validated only the accuracy of ventricular CSF and not TIV or TBV.
We present a fully-automated CT segmentation (CTSeg; https://github.com/NuroAI/CTSeg) method for brain segmentation and estimation of TBV and TIV from nonenhanced single-time-point head CT images by adapting SPM12. CTSeg was validated for brain segmentation and TBV and TIV estimation by comparing it with manual segmentation (n = 20). Additionally, we present a clinical application in which CTSeg is used to show TBV differences in Alzheimer disease (AD) (n = 116).
MATERIALS AND METHODS
Subjects
This study was reviewed and approved by the Geisinger Health System institutional review board. CT images were originally collected as part of patients’ routine clinical care but were fully de-identified. We created 2 datasets: 1) a manual segmentation dataset (n = 20, subjects free of brain abnormalities) and 2) an AD dataset (n = 167, subjects with and without a diagnosis of AD). Fifteen subjects with AD had catheters and were removed from further analysis. The AD dataset that was further analyzed consisted of 152 subjects.
Manual Segmentation Dataset.
A total of 20 subjects (mean age, 66 years; age range, 32–89 years; 10 women) were randomly selected for manual segmentation of the intracranial space and the brain parenchyma. These subjects were free of brain abnormalities and were unremarkable according to the radiology reports. Additionally, through visual inspection, we confirmed that the images were free of artifacts.
AD Dataset.
The initial cross-sectional AD dataset consisted of 62 subjects (mean age, 77 years; age range, 68–83 years; 41 women) with a diagnosis of AD and 90 controls (mean age, 78 years; age range, 68–83 years; 64 women) who did not have a diagnosis of AD. Subjects with AD and controls were selected on the basis of the International Classification of Disease, Ninth Revision (ICD-9-CM 331.0) codes.18 All CT images were free of artifacts, and the radiology reports of the images confirmed no acute pathologies or brain abnormalities. A retrospective evaluation indicated that the controls had undergone a CT scan following headaches or head injury. The CT images were acquired using multiple CT scanners, and details of the scanner models and imaging parameters are provided in On-line Table 1.
Manual Segmentation
Manual segmentation was performed by a trained operator using ITK-SNAP 3.6 (www.itksnap.org).19 The intracranial space was outlined according to the guidelines provided by Nordenskjöld et al.20 The segmented intracranial image was then used to segment the brain parenchyma by tracing the boundary between brain tissue and CSF.
Automated Brain Segmentation
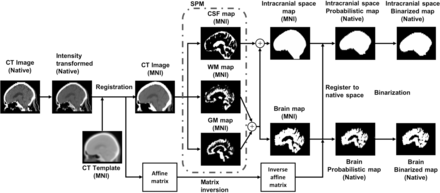
CTSeg (Fig 1) adapts the unified segmentation algorithm17 from SPM12 and uses a CT template for the initial affine registration step (see Methods: Automated Brain Segmentation in the On-line Appendix for a detailed explanation of the CTSeg pipeline). CTSeg creates probabilistic and binarized segmentation maps of the brain and the intracranial space. The binarized segmentation maps are obtained by thresholding the probabilistic maps using optimal thresholds (Optimal Threshold Selection in the On-line Appendix:).
CTSeg pipeline for intracranial space and brain parenchyma segmentation from head CT images. Within parentheses is the 3D coordinate space of the image. MNI indicates Montreal Neurological Institute.
Statistical Methods
The overlap between the automated and manual segmentation masks was measured using the Dice similarity index (DSI).21 TIV and TBV were obtained from the probabilistic maps, and the binary masks were obtained using CTSeg. For probabilistic maps, volumes were calculated by integrating the partial tissue volumes (tissue probability at the voxel × voxel volume) over all the voxels from the respective tissue maps. Volume estimates were calculated from binary masks by multiplying the number of masked voxels by the unit voxel volume.
Volumes estimated using CTSeg were compared with the manual estimates using scatterplots and the Pearson’s correlation coefficient. Systematic bias was assessed using Bland-Altman analysis,22 and percentage difference was calculated as a percentage of the manual estimates. The absolute agreement between automated and manual volumes was evaluated using the intraclass correlation coefficient (ICC) computed using 2-way ANOVA23 with fixed effects. The 95% confidence intervals of the ICC were computed using bootstrapping with 1000 iterations. The volumes were checked for normality using the Kolmogorov-Smirnov test.24 The TIV estimates of CTSeg were also compared with the state-of-the-art FSL Brain Extraction Tool for CT (BET; http://bit.ly/CTBET_BASH).13
CTSeg-estimated volumes from the images of age-matched subjects with AD and controls were used to compare brain atrophy between patients with AD and controls. Subjects with AD and controls were age-matched by minimizing the age difference using the MatchIt package25 in R (https://www.rdocumentation.org/packages/MatchIt/versions/3.0.2/topics/matchit).26 Previous studies have demonstrated that sex has no significant effect on TBV as a percentage of TIV (%TBV)27 because it is a normalized measure that accounts for the variability introduced by head size and sex.27,28 Therefore, subjects were not sex-matched because all our analyses were performed on %TBV. TBV versus TIV and %TBV versus age scatterplots were used to compare brain atrophy in patients with AD and controls. Linear regression models were used to determine the significance of age, sex, and AD diagnosis on %TBV. For the regression models, the Age × AD diagnosis interaction term was added to check whether the rate of brain volume loss was significantly different between patients with AD and controls. Additionally, we investigated the effect of TIV by modeling TBV, using TIV as a confounding factor in the linear models, as recommended in recent studies.20,29 TIV and sex were added in addition to Age and AD diagnosis while modeling TBV. Results with P < .05 are considered significant for all statistical analyses. Statistical analyses were performed using Python 2.7 (https://www.python.org/download/releases/2.7/), R 3.4.3 (http://www.r-project.org/), and Matlab 8.6.0. (MathWorks, Natick, Massachusetts).
RESULTS
Segmentation
CTSeg successfully segmented all 20 images from the manual segmentation dataset. The optimal image-intensity threshold obtained using a random selection of 10 training images was 0.2 for the brain mask and 0.0006 for the intracranial mask. These thresholds were robust when applied to the test set (On-line Fig 1).
Binary masks from CTSeg agreed well with the manual segmentation masks (the DSI was 0.94 ± 0.008 for brain and 0.98 ± 0.002 for the intracranial masks). The gyri and sulci in the superior slices of the brain were well-captured by CTSeg (On-line Fig 2).
Brain Volumetry
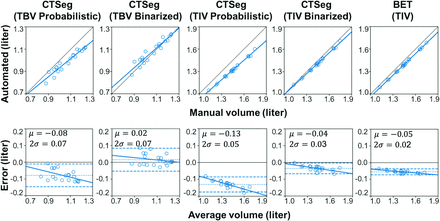
Comparison between automated and manual volume estimates is presented in Table 1. The binarized TBV and TIV estimates showed excellent agreement with the manual estimates (ICCs = 0.94 and 0.97, respectively), whereas the probabilistic estimates showed lower agreement (ICCs = 0.74 and 0.71 for TBV and TIV, respectively). TIV estimated using the BET also showed excellent agreement with manual TIV (ICC = 0.94) but was lower than binarized TIV from CTSeg. Binarized CTSeg also had the lowest bias in terms of the percentage difference (Table 1), and in the Bland-Altman plots (Fig 2) for both TIV (mean difference of −0.04L for CTSeg binarized versus −0.05L for BET and −0.13 for CTSeg probabilistic) and TBV (mean difference of 0.02L for CTSeg binarized versus −0.08L for CTSeg probabilistic for TBV). The pattern of the linear fit in the Bland-Altman plots showed that error increases with average volume and, therefore, head size for both estimates of CTSeg. However, the rate of increase was higher for probabilistic estimates than binarized estimates of CTSeg. The BET TIV estimate showed the lowest dependence of error on the average volume but showed larger bias than the binarized CTSeg TIV. Because the binarized CTSeg estimates showed better agreement with manual estimates, proceeding evaluations were made using only the binarized CTSeg method.
Comparison of automated TBV and TIV estimates with manual ground truth estimatesa
Scatterplots (upper row) of automated-versus-manual volume estimates and the linear fit between automated and manual volumes (thick line) and the line of equality (thin line). Bland-Altman plots (lower row) with automated-minus-manual volume differences on the y-axis and the average of automated and manual volumes on the x-axis. Mean difference ±2 SDs is represented by dotted and dashed horizontal lines, respectively.
Brain Volumetry in AD
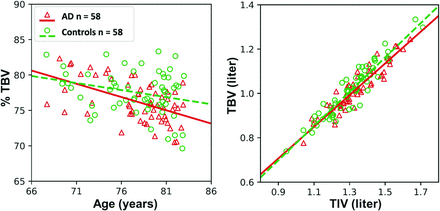
CTSeg was applied to the AD dataset containing 152 images. CTSeg successfully segmented 135 images (58 with AD and 77 controls) of 152 images (88%). Reasons for CTSeg failures are discussed in the next section. After we excluded CTSeg failures, 58 control subjects were optimally age-matched to 58 subjects with AD (a total of n = 116 subjects). A paired t test confirmed no significant age difference (P = .74) between the 2 groups after age-matching. Group comparisons were performed on binarized volumes estimated from the age-matched subjects. TBV and %TBV computed for AD and controls are presented in Fig 3. Linear fit to %TBV indicated a higher loss with age in the AD group than in controls. We observed significantly lower mean %TBV (P < .05) in the AD group (76.24 ± 2.87) compared to the control group (77.52 ± 3.05). A paired t test among %TBVs of the matched subjects also showed a significant difference (P < .05) between the two groups. The linear fit in the TBV-versus-TIV plot showed that the slope is lower for AD, suggesting a lower TBV-to-TIV ratio in subjects with AD. The results of the linear regression analysis are presented in On-line Table 2. Both age (P < .001) and AD diagnosis (P < .05) had a significant effect on %TBV. The Age × AD diagnosis term was insignificant as an interaction term in the linear model. Similar results were observed when TBV was modeled using sex and TIV as additional covariates. Age and AD diagnosis were significant when these variables were modeled as main effects. TIV had a significant contribution in all regression models. Results remained the same when sex was removed from the main effects model.
Scatterplot of %TBV (left) estimated using CTSeg versus age. Scatterplot of TBV versus TIV (right).
Segmentation Failures in the AD Dataset
CTSeg failed to produce acceptable segmentations for 4 of 62 AD images and 13 of 90 control images. Failures in the segmentation included segmentations of nonbrain regions like eyes as brain tissue or segmentation maps that did not resemble brain or intracranial space. On-line Table 3 summarizes the failure rate of CTSeg for different scanners. The overall failure rate was <15% across all the scanners.
DISCUSSION
TBV is an important measure for assessing brain atrophy in AD and other neurodegenerative diseases. Although CT is widely used in the clinical setting, segmentation methods to estimate TBV from head CT images are not available. We presented CTSeg, an automated head CT segmentation method, and validated the method by comparing it with manual segmentation.
TBV and TIV from binary CTSeg masks showed better agreement with manual estimates than the TBV and TIV estimates from probabilistic masks. This outcome was expected because the MR imaging–based default tissue probabilistic atlas map (TPM; https://www.fil.ion.ucl.ac.uk/spm/toolbox/TPM/) that we used did not model some of the anatomy present in CT images, and binarizing the masks by thresholding mitigated these errors. Additionally, the systematic bias in the TIV estimate using the binary masks was better than TIV estimated using the BET-based method by Muschelli et al.13
The utility of CTSeg was demonstrated in a cross-sectional dataset containing AD and control groups. We found that CTSeg-estimated volumes had a significant %TBV (P < .05) difference between the AD and control groups in a linear regression model with age, sex, and AD diagnosis as covariates. The sex of the subjects had no significant effect on the %TBV. This finding is in agreement with previous findings using MR images that used TIV to normalize global brain volumes.27,28 The average %TBV estimated from AD images was lower than that for matched controls. The statistical insignificance of Age × AD diagnosis interaction on %TBV can be attributed to the cross-sectional nature of our dataset. We expect that significant TBV group differences can be achieved if longitudinal head CT images of the same subjects are tracked. Furthermore, some of the %TBV variability may be due to not accounting for the duration from the actual onset of AD with respect to the time of the CT acquisition. Another factor that may have contributed to the %TBV is that our controls may have atrophy due to undiagnosed disorders. We expect to see higher group differences in TBV if ADs are compared with disorder free healthy controls. Additionally, when TBV is modeled with TIV as a confounding variable, we observed similar results compared with %TBV. TIV was a significant confounding variable in all the models. Sex was insignificant in all the models, suggesting that correction for TIV removes the structural differences between men and women; this finding agrees with previous ones using MR imaging–estimated volumes.29
Unlike MR imaging, the intensity of CT images is standardized and is a measure of radiation attenuation of the tissue. Therefore, we do not think that the scanner variability significantly affected our method. The standardized intensity in CT is, in fact, an advantage and makes the comparison of CT images across scanners easier, compared with MR images. Additionally, we expect the optimal thresholds of CTSeg to be widely applicable because SPM models the tissue intensities separately for each image. We confirmed the optimal threshold using two different approaches: random search and leave-one-out cross-validation. The high Dice similarity index in both approaches demonstrated the robustness of the optimal threshold. However, further validation on a larger dataset is required to verify the robustness of the threshold at different noise levels of CT images.
In CTSeg, we used a standard MR image–based TPM specific to an age range of 18–90 years for the segmentation.30 The CT template used for the initial registration was developed for an age range of 46–79 years.31 Although the age of the subjects used for this study was 67–89 years, we achieved good segmentation accuracy using the standard TPM and the CT template. However, if age-appropriate CT-based TPMs are used, we expect that segmentation accuracy would further improve. The TPM and the CT templates were created using images without brain abnormalities. Therefore, CTSeg assumes that the CT images to be segmented have brains that are free of large structural abnormalities like glioma, stroke, operations, and image artifacts due to beam-hardening and implants. However, CTSeg can be extended for applications for abnormal brain, like identifying lesions.32
CTSeg marginally overestimated TBV due to the misclassification of dura as brain in the superior slices of the image. This overestimation can be attributed to the low contrast-to-noise ratio among the soft tissues of CT images. Misclassification of the dura is a known problem even in the segmentation of T1-weighted MR images.33 TIV and TBV estimates from all automated methods tested in this study exhibited a linear dependence of error with head size. Binarizing the probabilistic maps using an optimal threshold slightly reduced this linear dependence to some extent, and this phenomenon can be attributed to several reasons: one reason may be the partial volume effect, in which a single voxel represents ≥2 tissues due to the finite spatial resolution of the image.34 The number of voxels at tissue boundaries increases with head size, thereby increasing the error in volume estimation due to the partial volume effect. The linear dependence of error and head size can also be attributed to errors in spatial registration and the allometric effect of the tissue priors. In the case of an intracranial mask, the optimal threshold was very low due to the influence of the high bone intensity (compared with CSF) on the partial volume effect for voxels near the bone-CSF interface.
We computed optimal thresholds for CT images with 5-mm image section thickness, which is the clinical standard for CT images. Because the partial volume effect increases with section thickness,35 thresholds may need to be derived independently for images with different section thicknesses. However, CT images reconstructed with smaller section thicknesses have a lower contrast-to-noise, which can lead to larger errors in the segmentation of brain tissue using CTSeg. Therefore, care should be taken when applying CTSeg to high-resolution images. On close visual inspection, we noted that some brain-CSF boundary regions were misclassified, especially in the left and right regions of the frontal lobe where the brain is closer to the skull and in regions between the brain hemispheres where the dura is present (On-line Fig 2). The misclassifications in intracranial maps (On-line Fig 3) were observed at the boundaries of the intracranial space in the superior and inferior slices, resulting in lower TIV estimates compared with manual segmentation. We also note that the binarized segmentation misclassified some parts of the eyes as the intracranial volume. This shortcoming can be corrected by registering a standard intracranial mask onto the binary intracranial mask obtained using CTSeg and excluding the voxels classified as TIV that are outside the registered standard intracranial mask.36
CONCLUSIONS
We present CTSeg to automatically estimate TBV and TIV from nonenhanced head CT images acquired for diagnostic purposes that were originally intended for visual evaluations by radiologists. We show that CTSeg can accurately estimate TBV and TIV. Application of CTSeg on CT images from subjects with AD and controls provides evidence that CTSeg can be used for detection and tracking of global brain atrophy in neurodegenerative diseases. AD does not have symptoms until the mild cognitive impairment stage, which occurs several years after the onset, and CTSeg may be used to track brain atrophy in these patients. In addition, CTSeg can be applied to clinical CT archives to develop normative brain volumes and to research studies involving neurodegenerative diseases that show global brain volume loss.
Acknowledgments
We thank the Geisinger Health System Foundation for the financial support.
Footnotes
This study was supported by the Geisinger Health System Foundation.
Disclosures: Stefi A. Baum—UNRELATED: Board Membership: Only in astrophysics, my primary research field; no money paid; Consultancy: only in astrophysics, my primary research field; money paid via the National Aeronautics and Space Administration. Employment: University of Manitoba, Comments: I am a professor of Physics and Astronomy and Dean of Science at the University of Manitoba; Grants/Grants Pending: only in Canada in my primary field of research, Astrophysics from the Natural Sciences and Engineering Research Council of Canada.* Maya Lichtenstein—UNRELATED: Grants/Grants Pending: Biohaven Pharmaceuticals/Alzheimer’s Disease Cooperative Study, Comments: Site Principal Investigator for the T2 Protect: Phase 2 Randomized Double Blind Placebo-Controlled Trial to Evaluate the Efficacy and Safety of BHV-4157 in Patients with Mild to Moderate Alzheimer's Disease.* *Money paid to the institution.
Indicates open access to non-subscribers at www.ajnr.org
References
- Received May 14, 2019.
- Accepted after revision November 20, 2019.
- © 2020 by American Journal of Neuroradiology