Abstract
BACKGROUND AND PURPOSE: Transcranial MR imaging–guided focused ultrasound is a promising novel technique to treat multiple disorders and diseases. Planning for transcranial MR imaging–guided focused ultrasound requires both a CT scan for skull density estimation and treatment-planning simulation and an MR imaging for target identification. It is desirable to simplify the clinical workflow of transcranial MR imaging–guided focused ultrasound treatment planning. The purpose of this study was to examine the feasibility of deep learning techniques to convert MR imaging ultrashort TE images directly to synthetic CT of the skull images for use in transcranial MR imaging–guided focused ultrasound treatment planning.
MATERIALS AND METHODS: The U-Net neural network was trained and tested on data obtained from 41 subjects (mean age, 66.4 ± 11.0 years; 15 women). The derived neural network model was evaluated using a k-fold cross-validation method. Derived acoustic properties were verified by comparing the whole skull-density ratio from deep learning synthesized CT of the skull with the reference CT of the skull. In addition, acoustic and temperature simulations were performed using the deep learning CT to predict the target temperature rise during transcranial MR imaging–guided focused ultrasound.
RESULTS: The derived deep learning model generates synthetic CT of the skull images that are highly comparable with the true CT of the skull images. Their intensities in Hounsfield units have a spatial correlation coefficient of 0.80 ± 0.08, a mean absolute error of 104.57 ± 21.33 HU, and a subject-wise correlation coefficient of 0.91. Furthermore, deep learning CT of the skull is reliable in the skull-density ratio estimation (r = 0.96). A simulation study showed that both the peak target temperatures and temperature distribution from deep learning CT are comparable with those of the reference CT.
CONCLUSIONS: The deep learning method can be used to simplify workflow associated with transcranial MR imaging–guided focused ultrasound.
ABBREVIATIONS:
- DL
- deep learning
- MAE
- mean absolute error
- SDR
- skull-density ratio
- tcMRgFUS
- transcranial MR imaging–guided focused ultrasound
- UTE
- ultrashort TE
Transcranial MR imaging–guided focused ultrasound (tcMRgFUS) is a promising novel technique for treating multiple disorders and diseases, including essential tremor,1 neuropathic pain,2 and Parkinson disease.3 During tcMRgFUS treatment, ultrasound energy is deposited from multiple ultrasound elements to a specific location in the brain to increase tissue temperature and ablate the targeted tissue. tcMRgFUS treatment-planning is usually performed in 3 steps: 1) CT images are acquired to estimate regional skull density and skull geometry and to estimate ultrasound attenuation during ultrasound wave propagation,1 2) MR images are acquired to identify the ablation target in the brain,1 and 3) the CT and MR images are fused to facilitate treatment-planning. Minimizing the steps involved to get to actual treatment can have a positive impact on clinical workflow. Here, we focus on the implications of minimizing patient burden by eliminating CT imaging (hence no radiation) and replacing it with synthesized CT of the skull based on ultrashort TE (UTE) images.
UTE MR imaging is an important technique for imaging short-T2 tissue components such as bone. A previous study has shown the feasibility of using UTE MR images for tcMRgFUS planning.4 The conversion of UTE MR images to CT intensity (also termed “synthetic CT”) is based on the inverse-log relationship between UTE and CT signal intensity.4,5
Deep learning (DL) with a convolutional neural network has recently led to breakthroughs in medical imaging fields such as image segmentation6 and computer-aided diagnosis.7 Domain transfer (eg, MR imaging to CT) is one of the fields in which DL has been applied recently with high accuracy and precision.8⇓⇓⇓-12 DL-based methods synthesize CT images either by classifying MR images into components (eg, tissue, air, and bone)10,12 or by directly converting MR imaging intensities into CT Hounsfield units.8,9,11 The established applications include MR imaging–based attenuation correction in MR imaging/PET10⇓-12 and treatment-planning for MR imaging–guided radiation therapy procedures.9 However, to our knowledge, DL methods have not been applied in the context of tcMRgFUS. In tcMRgFUS, our focus is skull CT intensity rather than the whole head as in the above procedures. By narrowing the focus area, we can potentially achieve higher accuracy in obtaining synthetic skull CT images from MR imaging.
The purpose of this study was to examine the feasibility of DL techniques to convert MR imaging dual-echo UTE images directly to synthetic CT of the skull images and assess its suitability for tcMRgFUS treatment-planning procedures.
MATERIALS AND METHODS
Study Participants
We retrospectively evaluated data obtained from 41 subjects (mean age, 66.4 ± 11.0 years; 15 women) who underwent the tcMRgFUS procedure and for whom both dual-echo UTE MR imaging and CT data were available. The study was approved by the institutional review board (University of Maryland at Baltimore).
Image Acquisition and Data Preprocessing
MR brain images were acquired on a 3T system (Magnetom Trio; Siemens) using a 12-channel head coil. A prototype 3D radial UTE sequence with 2 TEs was acquired in all subjects.13 Imaging parameters were the following: 60,000 radial views, TE1/TE2 = 0.07 /4 ms, TR= 5 ms, flip angle = 5°, matrix size = 192 × 192 × 192, spatial resolution = 1.3 × 1.3 × 1.3 mm3, scan time = 5 minutes.13,14 CT images at 120 kV were acquired using a 64-section CT scanner (Brilliance 64; Philips Healthcare), with a reconstructed matrix size = 512 × 512 and resolution = 0.48 × 0.48 × 1 mm3. A C-filter (Philips Healthcare), a Hounsfield unit–preserving sharp ramp filter, was applied to all images.
UTE images were corrected for signal inhomogeneity with nonparametric nonuniform intensity normalization bias correction using Medical Image Processing, Analysis, and Visualization (National Institutes of Health).15 Both UTE volumes (TE1 and TE2) for each subject were normalized by the tissue signal of the TE1 image to account for signal variation across subjects. CT images from each subject were registered and resampled to the corresponding UTE images using the FMRIB Linear Image Registration Tool (FLIRT; https://fsl.fmrib.ox.ac.uk/fsl/fslwiki/FLIRT) using a normalized mutual information cost function.16 Finally, CT of the skull images were derived by segmenting the registered CT images using automatic image thresholding with the Otsu method.17 The same threshold value was applied to the DL synthetic CT (DL-CT) data. Only the skull from the superior slices relevant for the tcMRgFUS procedure from both UTE and CT was included. CT-UTE registration results were evaluated by visual inspection. One subject was excluded due to failed registration, leading to 40 subjects in total (mean age, 66.5 ± 11.2 years; 15 women).
Deep Learning Model and Neural Network Training
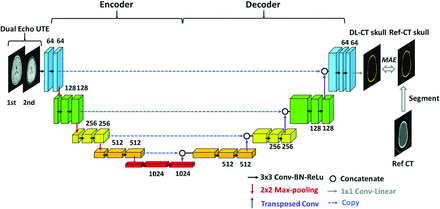
A schematic diagram of the deep learning model architecture based on U-Net convolutional neural network (https://lmb.informatik.uni-freiburg.de/people/ronneber/u-net/)18 is illustrated in Fig 1. It consists of 2 pathways in opposite directions: encoding and decoding. The encoding pathway extracts features of the input images, while the decoding pathway has the opposite direction, restoring these features.
Schema of the employed deep learning architecture based on the widely used U-Net convolutional neural network consisting of encoding and decoding pathways. Dual-echo UTE images were used as the input for the network. Reference CT of the skull was segmented from the reference CT and was used as the predication target. The difference between output of the network, DL synthetic CT of the skull, and reference CT of the skull was minimized using MAE loss function. Drop-out regularization (rate = 0.5) was applied connecting the encoder and decoder. BN indicates batch normalization; ReLu, rectified linear unit; Conv, convolutional layer; Ref-CT, reference CT.
Dual-echo UTE images were used as the input to the neural network, with each echo as a separate input channel to the network. Reference CT of the skull images was used as the prediction target. Output of the network is the DL-CT of the skull.
UTE-CT image pairs from 32 subjects were selected as the training dataset, and the other 8, as the testing dataset. The neural network was defined, trained, and tested using Keras with Tensorflow backend (https://www.tensorflow.org) with a Tesla K40C GPU (memory of 12GB). Mean absolute error (MAE) between DL-CT of the skull and the reference CT of the skull was minimized using the ADAM algorithm, derived from adaptive moment estimation.19 The training was performed with 100 epochs, and the learning rate was 0.001. Training of the model took approximately 6 hours.
Evaluation of Model Performance
The performance of the neural network model was evaluated using the 5-fold cross-validation method. The 40 subjects were randomly divided into 5 groups, each with 8 subjects. Each time 1 group was held as a testing dataset, the other 4 served as the training datasets; thus, the model was trained 5 separate times. Testing results from all 40 subjects were used to evaluate the performance of the model. The following 4 metrics were used to compare the DL-CT from the testing datasets with the reference CT of the skull images: 1) Dice coefficient to evaluate the similarity between the 2 sets of images; 2) a voxelwise spatial correlation coefficient between the 2 methods considering all the voxel intensities (in Hounsfield units); 3) average of absolute differences between the 2 methods for voxels within skull region; and 4) global CT Hounsfield unit values for each subject by averaging all the voxels within the skull region. Metrics using a conventional method4 were also derived to enable comparison with the DL method. Note that only testing datasets (n = 40) from cross-validation results were used for evaluation and the following validation/simulation.
Skull Density Ratio Validation
To further evaluate the accuracy of DL-CT-derived skull properties, we also calculated the regional skull thickness and the skull-density ratio (SDR) based on each of the 1024 ultrasound rays on 40 subjects. We validated our model by comparing the whole SDR from DL-CT of the skulls with reference CT of the skulls. The SDR is calculated as the ratio of the minimum over the maximum intensity (Hounsfield unit) along the skull path of ultrasonic waves from each of the 1024 transducers, and the whole-skull SDR is the average of all SDRs. Whole-skull SDR > 0.45 is the eligibility criterion for tcMRgFUS treatment for efficient sonication.20
Acoustic and Temperature Simulation
A key aspect of the tcMRgFUS is the guidance obtained from MR thermometry maps during the procedure, eg, to predict the target temperature rise. We therefore compared acoustic and temperature fields from the simulation using both CT and DL-CT images on 8 test subjects. The acoustic fields within the head were simulated using a 3D finite-difference algorithm, which aims to solve the full Westervelt equation.21 The acoustic properties of the skull were derived from CT images of the subjects. Temperature rise was estimated using the inhomogeneous Pennes equation22 of heat conduction with the calculated acoustic intensity field as input. Both acoustic and temperature simulations were performed with the target at the center of the anterior/posterior commissure line for each subject at a spatial resolution of UTE scans. Temperature elevations at the focal spots caused by a 16-second, 1000-W sonication were simulated for both reference CT and DL-CT images, assuming that the base temperature of the brain was 37°C.
RESULTS
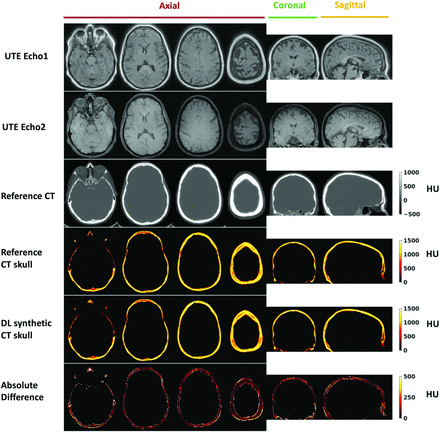
Figure 2 shows the DL-CT of the skull images in comparison with the reference CT of the skull images from a representative test subject. The difference images show minimal discrepancy, demonstrating that the trained DL network has successfully learned the mapping from dual-echo UTE images to CT of the skull-intensity images. The On-line Figure shows the DL model loss (MAE) as a function of the epoch number for both the training and testing datasets from a representative cross-validation run.
Deep learning results from 1 representative testing subject (55 years of age, female). From top to bottom: UTE echo 1 and echo 2 images, reference CT, segmented reference CT of the skull, DL synthetic CT of the skull, and the absolute difference between the 2. For this subject, the Dice coefficient for skull masks between DL synthetic CT and the reference CT is 0.92, and the mean absolute difference is 96.27 HU.
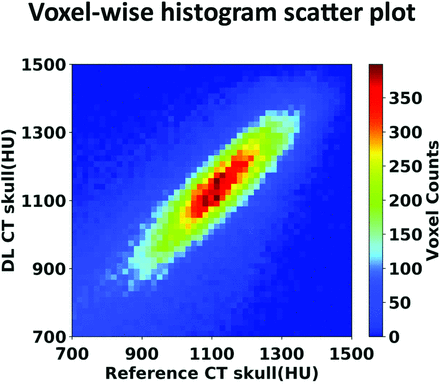
The signal intensities between the 2 CT scans are highly correlated (r = 0.80) as shown in the voxelwise 2D histogram (Fig 3), demonstrating that DL can accurately predict the spatial variation across regions within the skull. The Table summarizes various metrics estimating the performance of the DL model from all 5 runs of the cross-validation process, along with the average from all 40 testing datasets. As shown in the Table, model performance is comparable between different runs in the cross-validation process. As a comparison, the MAE and spatial correlation coefficient from the conventional method4 were 0.37 ± 0.09 and 432.19 ± 46.61 HU, performances significantly poorer than our proposed deep learning method.
Voxelwise 2D histogram scatterplot between the reference skull CT intensity and the DL synthetic skull CT signal intensity in Hounsfield units within skull. The correlation coefficient is r = 0.80 (same testing subject as in Fig 2). Color bar represents the voxel count.
Various metrics (mean ± standard deviation) showing the performance of the deep learning model from all 5 runs of the cross-validation processa
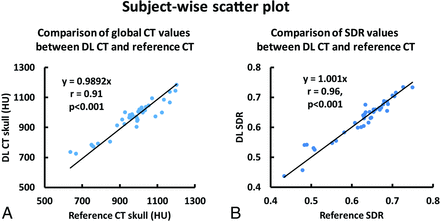
Subject-wise scatterplots of average CT values and SDR values are shown in Fig 4. A strong correlation between the DL-CT and the reference CT Hounsfield unit values (r = 0.91, P < .001) was observed, demonstrating that the derived model can predict the global intensity of the skull accurately. The high whole-skull SDR correlation (r = 0.96, P < .001) between DL-CT and the reference CT suggests a strong potential for the use of DL-CT images for treatment-planning in tcMRgFUS.
A, Association of average CT Hounsfield unit values between the DL synthetic CT of the skull and the reference CT of the skull for all 40 testing subjects from cross-validation. Each dot represents 1 subject. B, The relationship between SDR values determined from the reference CT and DL synthetic CT from all 40 subjects. Each dot represents 1 subject.
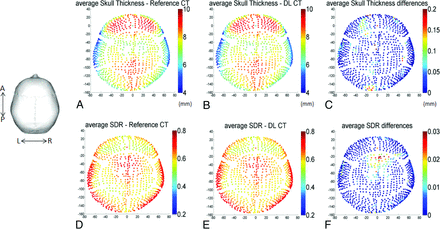
Figure 5 shows averaged regional skull thickness maps Fig 5A, -B and averaged regional SDR maps (Figure 5D, -E) based on the reference CT and DL-CT images from all 40 test subjects. The errors in skull thickness measurement at any given entry were <0.2 mm (2%), averaged at 0.03 mm (0.3%) (Fig 5C). The maximum error for SDR calculation across the 1024 entries was found to be about 0.03 (4%) and averaged less than 0.01 (1.3%) (Fig 5F).
A and B, The calculated average skull thickness map from the reference CT and the DL synthetic CT images, respectively, from all 40 testing subjects. C, The differences between A and B with a maximum thickness difference of 0.2 mm (2% error) and the average error of 0.03 mm (0.3%). Note that regional maps are based on the entries of the 1024 ultrasound beams from the ExAblate system (InSightec). D and E, The calculated average SDR map based on the reference CT and DL synthetic CT images from all 40 subjects. F, The differences between D and E with a maximum SDR difference of 0.03 (4% error) and the average error of 1024 entries was <0.01 (1.3%).
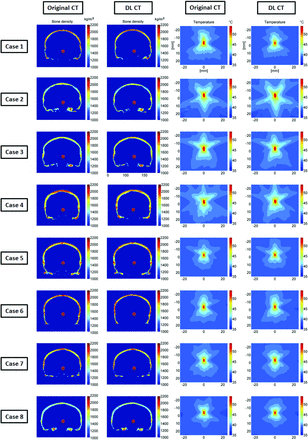
Comparisons between calculated bone density and the simulated temperature rise results are shown in Fig 6 for 8 representative test subjects. The estimated voxelwise average bone density difference within the skull for all subjects was 50 kg/m3, indicating less than an average of 2.3% error for UTE-derived acoustic properties compared with the reference CT images (columns 1 and 2). Simulated temperature patterns are very similar for all 8 cases (columns 3 and 4), indicating that the skull shape and thickness of DL-CT are highly comparable with the reference CT. The differences of simulated peak temperature rise based on original and DL synthetic CT images for all 8 subjects are well within the errors that one might expect from the simulation.
Comparison of calculated bone density and the simulated temperature rise. The first and second columns show the calculated bone density map using the reference CT and DL synthetic CT images on 8 representative testing cases, in which the red dots are the assigned focal targets. In the third and fourth columns, the simulated temperature elevations at the focal spots caused by a 16-second, 1000-W sonication are compared between reference CT and DL synthetic CT on a base brain temperature of 37°C. The simulated peak temperature rise values based on original and DL synthetic CT images for all 8 subjects are the following: case 1: 55.3°C and 54.2°C (6.0% error on temperature rise), case 2: 59.0°C and 58.9°C (0.5% error), case 3: 57.6°C and 55.9°C (8.2% error), case 4: 57.5°C and 59.1°C (7.8% error), case 5: 54.3°C and 54.6°C (0.6% error), case 6: 55.5°C and 56.0°C (0.9% error), case 7: 54.5°C and 55.1°C (1.1% error), case 8: 53.5°C and 54.5°C (1.9% error), respectively. These errors are well within the errors that one might expect from the simulation.
DISCUSSION
In this study, we examined the feasibility of leveraging deep learning techniques to convert MR imaging dual-echo UTE images directly to CT of the skull images and to assess the applicability of such derived images to replace real CT of the skull images during tcMRgFUS procedures. The proposed neural network is capable of accurately predicting Hounsfield unit intensity values within the skull. Various metrics were used to validate the DL model, and they all demonstrated excellent correspondence between DL-CT of the skull images and the reference CT of the skull images. Furthermore, the acoustic properties as measured using the SDR and temperature simulation suggest that DL-CT images can be used to predict target temperature rise. Our proposed DL model has the potential to eliminate CT during tcMRgFUS planning, thereby simplifying clinical workflow and reducing the number of patient visits and overall procedural costs, while eliminating patient exposure to ionizing radiation. To our knowledge, this is the first study to apply deep learning in synthesizing CT of the skull images with MR imaging UTE images for tcMRgFUS treatment planning.
Several previous studies have reported methods to derive CT information from MR imaging.9⇓⇓-12,23 Multiple DL-based CT synthesis studies have reported CT bone segmentation with Dice coefficient values ranging from 0.80 to 0.88.9⇓⇓-12,23 In our study, the proposed model shows excellent performance in estimating CT-equivalent skull with a Dice score of 0.91 ± 0.03 for all 40 testing datasets. The moderately higher level of performance over other previous methods may be because only a limited amount of the skull was taken into consideration due to its relevance for tcMRgFUS, whereas previous methods also included brain tissue.15⇓⇓⇓⇓⇓-21 Furthermore, our model predicted CT intensity with a high voxelwise correlation (0.80 ± 0.08) and low MAE (104.57 ± 21.33 HU) for all 40 subjects compared with the reference CT of the skull. One study4 reported an MAE between the reference CT and UTE-generated synthetic CT of 202 HU in the context of tcMRgFUS. Applying this method4 to our dataset, we observed a higher MAE of 432.19 ± 46.61 HU. The discrepancy may be due to differences of the subject cohort and skull mask delineation. Another study reported an MAE of 174 ± 29 HU within the skull.9 Compared with these studies, our results represent a marked improvement over existing methods.
While this is the first report demonstrating the feasibility of applying deep learning in tcMRgFUS, our proposed framework can be further improved in a few ways: First, the DL field is rapidly evolving, and newer state-of-the-art techniques continue to emerge. The inputs to the implemented 2D U-Net neural network in our case were individual 2D dual-echo UTE images to generate 2D CT of the skull images as output. It is highly possible that a 2.5D or 3D U-Net may further minimize these errors because these approaches use context information for training. However, note that the 3D U-Net requires significantly more memory resources than the 2D U-Net. Additionally, alternative loss formulations or combinations may be considered (eg, an adversarial component to the loss function to maximize the realistic appearance of the generated output). Given the relatively small dataset and the fact that the MR imaging-to-CT mapping task does not require full-FOV MR imaging, an alternative approach may be to train a patch-wise classifier (same encoder-decoder architecture, simply smaller). Not only will the model be more compact, it will likely be more regularized and more generalizable to edge cases (eg, craniotomy).
This study has several limitations. One limitation is that the average age of our patient cohort is relatively high (66.5 ± 11.2 years of age). This might limit the usage of our model in younger cohorts or pediatric populations due to bone density variations. Incorporating data from younger subjects into our training data can address this issue. Another limitation is our relatively small sample size for the deep learning study and the lack of an independent test dataset. More datasets will certainly improve the performance of the model and allow better generalization of our model. Additionally, our CT of the skull synthesis was based on MR imaging UTE images, which have relatively low spatial resolution compared with CT (1.33 versus 0.44 mm in-plane resolution). This resolution discrepancy might affect the accuracy of our model in predicting the skull mask and Hounsfield unit values. To address this issue, high-resolution UTE 3D images are needed using advanced parallel imaging, compressed sensing, or even DL-based undersampling/reconstruction to further reduce the scan time while preserving enough information for CT synthesis. Finally, we will investigate the effect of data augmentation on larger datasets in detail and use an advanced deep learning model such as the Generative Adversarial Network (https://github.com/goodfeli/adversarial) to further improve our model in a future study.
CONCLUSIONS
We examined the feasibility of using DL-based models to automatically convert dual-echo UTE images to synthetic CT of the skull images. Validation of our model was performed using various metrics (Dice coefficient, voxelwise correlation, MAE, global CT value) and by comparing both global and regional SDRs derived from DL and the reference CT. Additionally, temperature simulation results suggest that DL-CT images can be used to predict target temperature rise. Our proposed DL model shows promise for replacing the CT scan with UTE images during tcMRgFUS planning, thereby simplifying workflow.
Footnotes
P. Su and S. Guo contributed equally to this work.
Disclosures: Pan Su—RELATED: Other: employment: Siemens Healthineers. Florian Maier—UNRELATED: Employment: Siemens Healthcare GmbH; Stock/Stock Options: Siemens Healthineers AG, Comments: I am a shareholder. Himanshu Bhat—RELATED: Other: Siemens Healthineers, Comments: employment. Dheeraj Gandhi—UNRELATED: Board Membership: Insightec*; Grants/Grants Pending: MicroVention, Focused Ultrasound Foundation*; Royalties: Cambridge Press. *Money paid to the institution.
References
- Received March 30, 2020.
- Accepted after revision July 5, 2020.
- © 2020 by American Journal of Neuroradiology