Abstract
BACKGROUND AND PURPOSE: Currently, accurate and reproducible spinal cord GM segmentation remains challenging and a noninvasive broadly accepted reference standard for spinal cord GM measurements is still a matter of ongoing discussion. Our aim was to assess the reproducibility and accuracy of cervical spinal cord GM and WM cross-sectional area measurements using averaged magnetization inversion recovery acquisitions images and a fully-automatic postprocessing segmentation algorithm.
MATERIALS AND METHODS: The cervical spinal cord of 24 healthy subjects (14 women; mean age, 40 ± 11 years) was scanned in a test-retest fashion on a 3T MR imaging system. Twelve axial averaged magnetization inversion recovery acquisitions slices were acquired over a 48-mm cord segment. GM and WM were both manually segmented by 2 experienced readers and compared with an automatic variational segmentation algorithm with a shape prior modified for 3D data with a slice similarity prior. Precision and accuracy of the automatic method were evaluated using coefficients of variation and Dice similarity coefficients.
RESULTS: The mean GM area was 17.20 ± 2.28 mm2 and the mean WM area was 72.71 ± 7.55 mm2 using the automatic method. Reproducibility was high for both methods, while being better for the automatic approach (all mean automatic coefficients of variation, ≤4.77%; all differences, P < .001). The accuracy of the automatic method compared with the manual reference standard was excellent (mean Dice similarity coefficients: 0.86 ± 0.04 for GM and 0.90 ± 0.03 for WM). The automatic approach demonstrated similar coefficients of variation between intra- and intersession reproducibility as well as among all acquired spinal cord slices.
CONCLUSIONS: Our novel approach including the averaged magnetization inversion recovery acquisitions sequence and a fully-automated postprocessing segmentation algorithm demonstrated an accurate and reproducible spinal cord GM and WM segmentation. This pipeline is promising for both the exploration of longitudinal structural GM changes and application in clinical settings in disorders affecting the spinal cord.
ABBREVIATIONS:
- AMIRA
- averaged magnetization inversion recovery acquisitions
- CV
- coefficient of variation
- DSC
- Dice similarity coefficient
- HD
- Hausdorff distance
- SC
- spinal cord
The human spinal cord (SC) can be affected by numerous neurologic disorders of variable pathophysiology (eg, genetic, inflammatory, demyelinating, degenerative, and so forth),1,2 and MR imaging is a valuable part of the diagnostic work-up in patients with suspected intramedullary pathology.3,4 SC gray matter and white matter can be involved to a various extent not only among different SC disorders but also among patients with the same disease (eg, multiple sclerosis, amyotrophic lateral sclerosis).5,6 Hence, quantification of SC compartments may add to our understanding of SC pathology5,6 and hopefully help in the management of individual patients in the future.
However, the SC presents additional challenges for MR imaging. The SC is surrounded by a number of different tissue types, including CSF, bone, and air. These create considerable signal inhomogeneities along this thin, elongated structure.3,4,7,8 As a result, conventional SC MR imaging was—until recently—not able to differentiate sufficiently among SC GM, WM, and CSF. In the past, there were first attempts toward this differentiation using a series of acquisition approaches.9⇓⇓–12 More recently an averaged magnetization inversion recovery acquisitions (AMIRA) sequence was proposed, delivering a notable SC GM/WM contrast while maintaining short acquisition times at the same time.13 The latter is especially important for imaging small-sized structures (like the SC GM/WM) in patients with disabilities having a short time window in which they can lie still.
Moreover, accurate SC GM segmentation remains challenging. First, manual approaches demonstrated the feasibility of distinguishing between WM and GM.9 However, manual approaches require a considerable amount of time, are prone to error, and demonstrate significant interobserver and intraobserver variability. As a result of improvements in image quality and postprocessing techniques, the first fully automatic SC GM segmentation methods were established in the past few years.14⇓⇓–17 These methods have deployed atlas-based GM segmentation algorithms, which may, however, lead to misestimations or segmentation errors, especially in case of pathology, image artifacts, or large between-individual anatomic variations.18,19 A noninvasive broadly accepted reference standard for accurate and reproducible SC GM measurements is still a matter of ongoing discussion.
In this study, we validate a fully automatic method for SC GM and WM segmentation in terms of its reproducibility and accuracy in segmenting the cervical SC of healthy controls against a manual segmentation. The proposed approach used a variational segmentation algorithm with a shape prior,20 modified for 3D data with a slice similarity prior on AMIRA images.
Materials and Methods
Subjects and MR Imaging Acquisition
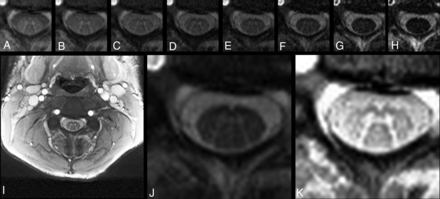
Twenty-four healthy subjects (14 women; mean age, 40 ± 11 years) were scanned in a test-retest fashion on a 3T whole-body MR imaging system (Magnetom Prisma; Siemens, Erlangen, Germany). All subjects provided written consent. Experimental procedures conformed to the Declaration of Helsinki, and the study protocol was approved by the local ethics committee. We acquired 12 axial AMIRA images13 (FOV = 128 × 128 mm2, slice thickness = 8 mm, slice overlap = 4 mm, in-plane resolution = 0.67 × 0.67 mm2, TEbalanced steady-state free precession = 2.14 ms, TRbalanced steady-state free precession = 5.13 ms, signal averaging = 1, acquisition time = 51 seconds per slice) over a 48-mm cervical SC segment, extending approximately from the C2–C5 vertebral level.13 The most rostrally acquired slice was placed with its lower surface adjacent to the most rostral surface of the C2/C3 intervertebral disc. For precise positioning of each individual slice and its orthogonal angulation to the course of the SC, a strongly T2-weighted TSE with high contrast between CSF and SC was used as a reference. For each slice, the AMIRA approach acquired 8 images of considerably different tissue contrast among GM, WM, and CSF with effective TI = 97.1, 158.7, 220.2, 281.8, 343.3, 404.9, 466.5, 528.0 ms. Averaging the first 5 images enhances the GM/WM contrast-to-noise ratio, whereas averaging the last 3 images clearly improves the WM/CSF contrast-to-noise ratio (Fig 1). For more details on the AMIRA sequence, please see Weigel and Bieri, 2018.13
Exemplary axial AMIRA slice of 1 representative volunteer at the C4 level. A–H, Eight images of different tissue contrast acquired by the AMIRA sequence, shown in chronologic order from lowest-to-highest TI. I, Average image from A to E in full view, which delivers a high contrast-to-noise-ratio for GM/WM. J, Average image from F to H, which delivers a high contrast-to-noise ratio for SC/CSF. K, Same average image as in I but histogram-equalized and zoomed.
Each subject was scanned 3 times in 1 MR imaging session. The first 2 scans were performed in a back-to-back fashion without repositioning to allow intrasession comparisons. The third scan was obtained after patient repositioning to allow intersession comparisons.
All scans underwent basic preprocessing including 2D and 3D correction for field inhomogeneities using the scanner software before segmentation. To minimize numeric errors of the validation metrics, we performed a 5-fold in-slice upsampling of the slices using the Lanczos-3 interpolation kernel.
SC Segmentation
As proposed in a previous study,20 a variational segmentation approach based on the continuous min-cut max-flow framework was used, which includes total variation regularization to segment WM and GM. The min-cut max-flow capacity functions are modeled using edge, region, and prior information as well as an appearance model built from manual segmentations. Aiming for high accuracy, the proposed approach prefers intensities of the actual image and tries to include prior information as little as possible, which regularizes for higher precision. Compared with the previous study,20 we added a slice similarity prior,21 included all inversion images of one slice (Fig 1) into the calculation of the max-flow capacity functions, and improved the adaptation of the appearance model with posterior models that reconstruct the most likely appearance only based on well segmented pixels.22
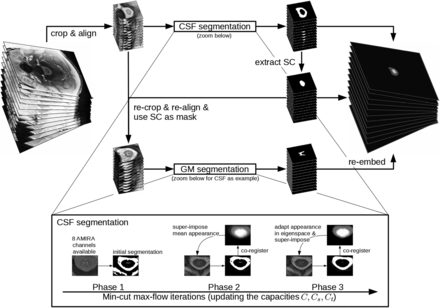
As a first step of the algorithm to align the 12 slices, the images are center-cropped and slice-wise successively coregistered rostral to caudal using translations in pixel-size steps to prevent further interpolation. Then, the algorithm automatically locates and delineates the ring-shaped CSF from its surroundings and extracts the cross-sectional SC surface. Finally, it uses the previously segmented SC surface as a mask for GM/WM differentiation. An illustration of the algorithm is shown in Fig 2. Segmentations were achieved in a leave-one-subject-out cross-validation—that is, with the currently segmented subject being left out in the appearance model used.
Flow chart of the automatic segmentation pipeline. As a first step of the algorithm to align the 12 slices, the images are center-cropped and slice-wise successively coregistered rostral to caudal using translations in pixel-size steps to prevent further interpolation. Then, the algorithm automatically locates and delineates the ring-shaped CSF from its surroundings and extracts the cross-sectional SC surface. Finally, it uses the previously segmented SC surface as a mask for GM/WM differentiation. The iterative steps of CSF segmentation are shown as a zoomed-in view. GM segmentation uses essentially the same steps and is thus not shown in detail.
The segmentation algorithm was implemented in Matlab (MathWorks, Natick, Massachusetts). Processing time on a Xeon CPU E5–2620 v3 @ 2.40GHz (Intel, Santa Clara, California) is around 1 minute for each segmentation step (CSF-SC and WM-GM segmentation), and fewer than 8 GB of RAM is used to segment a stack of 12 slices. Code is available on https://github.com/neonroehre/AJNR2019.
Two experienced raters (C.T. and A.A.) were involved in the manual segmentations. Both raters had >4 years' experience in neuroimaging research, including SC volumetric studies. In a first step, segmentations were conducted on the average of the last 3 AMIRA images for the total SC cross-sectional area. Using the already delineated total SC masks, we then performed manual segmentations of the GM and WM cross-sectional areas on the average of the first 5 AMIRA images (On-line Figure). C.T. segmented all images once. These results were further applied as a “manual reference standard.” C.T. also conducted a second “run” of 60 randomly selected slices to assess intrarater comparisons. This second run was conducted with slightly different contrast adjustments than the first to evaluate the robustness of intrarater manual segmentation. A.A. segmented all images of the first scan of all 24 healthy controls to allow interrater comparisons.
To evaluate the performance of our method on SC slices, in which the fully automatic approach failed (in total 12% of acquired slices, see also Results), we applied a semiautomatic approach as follows: The SC/CSF boundaries were segmented manually (manual reference standard) and segmentation of the GM and WM was then performed using the fully-automatic approach described above, given the manual total SC masks.
To compare our automatic method with currently available algorithms, we tested the iterative nonlocal STAPLE algorithm23 on our AMIRA images using the algorithm in the SCFusion_Demo package (https://www.nitrc.org/frs/download.php/7666/scfusion_demo.zip). Asman et al23 used atlases consisting of SC-GM-WM contrast images and SC-GM-WM manual reference standard segmentations, which are rigidly registered to the target slice and fused together with the most fitting manual reference standard segmentation as an estimation of the targeted segmentation. We built our own atlases and tested the iterative nonlocal STAPLE in a leave-one-subject-out fashion.
Statistical Analysis
Intra- and intersession and intra- and interrater reproducibility of the 2 approaches were evaluated using coefficients of variation (CVs), Dice similarity coefficients (DSCs), and Hausdorff distances (HDs). The accuracy of the automatic method compared with the manual reference standard was evaluated using the DSC and HD. CVs between the 2 masks A and B were calculated with the following formula:
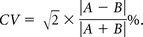 DSCs were calculated as follows:
DSCs were calculated as follows:
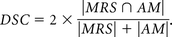 HDs were calculated as follows:
HDs were calculated as follows:
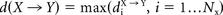
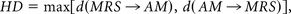 where d is the Euclidean distance between voxels x and y, MRS is the manual reference standard and AM the automatic method.
where d is the Euclidean distance between voxels x and y, MRS is the manual reference standard and AM the automatic method.
Because of non-normally distributed data, we performed a square root transformation of the CV, a cubic transformation of the DSC, and a logarithmic transformation of the HD before conducting all t test analyses and MANOVA. Two-paired t tests were performed for the following comparisons after controlling for normal data distribution: 1) manual segmentation versus automatic method reproducibility, and 2) manual segmentation versus automatic method total SC, WM, and GM cross-sectional area. For the automatic method, differences in measures of reproducibility and accuracy between intrasession and intersession; among GM, WM, and total SC; and among the axial slice levels (1–12) were investigated using MANOVA. Additional Tukey post hoc tests were conducted when applicable.
Results
In total, 864 slices were acquired from 24 volunteers with 12 slices per scan, and each scan was performed a total of 3 times for each subject. Of 864 acquired axial SC slices, 9 were excluded from further analysis because of severe imaging artifacts. The automatic method successfully segmented 88% (752 slices) of all remaining slices. Because of imaging artifacts, localization problems, or posterior gaps of the CSF, 8% of all slice-wise SC segmentations and 4% of GM segmentations would have needed further manual interventions and thus were excluded from the reproducibility analysis.
Cross-Sectional SC Measurements
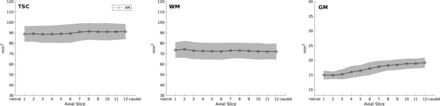
The mean total SC area was 89.98 ± 7.88 mm2, the mean WM area was 72.71 ± 7.55 mm2, and the mean GM area was 17.20 ± 2.28 mm2 as measured by the automatic method. Compared with the manual reference standard, the automatic method delivered significantly higher total SC and WM area as well as significantly lower GM area (86.88 ± 11.87, 69.18 ± 10.16, and 17.77 ± 3.05 mm2, respectively; all, P < .001). Cross-sectional areas per slice of the automatic method are shown in Fig 3.
Cross-sectional areas of total spinal cord, white matter, and gray matter per axial slice as measured by automatic segmentations. Notice the slight increase of total spinal cord (TSC) and the marked GM cross-sectional area increase caudally, which corresponds to the cervical SC enlargement. The light gray area depicts the limits of ±1 SD.
Reproducibility
Measurements of intra- and intersession and intra- and interrater reproducibility are shown in the On-line Table. Reproducibility of SC GM and WM is also depicted per slice in Fig 4. All mean CVs of the automatic method were ≤4.77%, and the mean DSC was ≥0.88 between scans and raters. The latter was significantly better than for the manual segmentation (all, P < .001).
Comparison between the reproducibility of manual and automatic measurements (AM) of spinal cord gray matter and white matter per axial slice. Intrasession and intersession reproducibility is assessed in terms of Dice coefficients (graphics on the left) and coefficients of variation (graphics on the right). Manual and automatic intersession reproducibility is shown in dark gray, whereas manual and automatic intrasession reproducibility is shown in light gray. Error bars display mean values ± 0.2 SDs.
By means of MANOVA with DSC, HD, and CV as multivariate outcomes, a significant difference between intra- and intersession reproducibility for total SC, WM, and GM using the automatic method was shown (P < .001 for all 3 models). However, CVs differed only for WM and total SC (P < .05 and P < .001, respectively), but not for GM. In our automatic method, intra- and intersession reproducibility was significantly decreased in the order total SC→WM→GM (all P < .001), as shown by MANOVA and post hoc tests. No difference was found in intra- and intersession reproducibility among slices for GM, but a significant decrease was found for WM (both P < .001) and total SC (P < .05 and P < .001, respectively) in caudal slices, as shown by MANOVA. However, CVs were similar for all SC metrics among all slices.
Accuracy
Measurements of accuracy of our fully automatic method compared with a manual reference standard are shown in detail in the Table and are also shown per slice in Fig 5. The automatic method showed a mean DSC of ≥0.86 in all SC metrics. Accuracy was significantly decreased in the order total SC→WM→GM (all P < .001), as shown by MANOVA and post hoc tests. In MANOVA, accuracy was lower for GM (P < .05) and total SC (P < .001) in caudal slices, but not for WM. However, the DSC was similar among acquired slices for total SC.
SC GM, WM, and TSC accuracy of automatic and semiautomatic segmentations against the manual reference standard segmentationsa
Accuracy measurements in terms of Dice coefficients (graphics on the left) and Hausdorff distances (graphics on the right) of white matter and gray matter per slice. Overlaid boxplots display median values as well as 25th and 75th percentile values. Gray areas depict the mean standard error values ± 1 SD.
Measurements of accuracy of the initially discarded SC slices (12% of all acquired AMIRA slices) analyzed in a semiautomatic fashion are also shown in the Table. The semiautomatic approach showed a mean DSC of ≥0.83 in both GM and WM. When we compared it with the fully automatic approach on the initially nondiscarded SC slices, a statistically significant accuracy decrease was observed in the semiautomatic approach (both P < .001).
Comparison with the Iterative Nonlocal STAPLE Algorithm
In comparison with the original study23 performed on T2* MR images, the application of the iterative nonlocal STAPLE algorithm in our AMIRA images showed a higher accuracy. Mean DSC and HD for the total SC, GM, and WM were as follows—total SC: mean DSC = 0.93 ± 0.03 (median, 0.94), mean HD = 0.96 ± 0.39 mm (median, 0.84 mm); GM: mean DSC = 0.80 ± 0.06 (median, 0.82), mean HD = 1.09 ± 0.42 mm (median, 1.04 mm); WM: mean DSC = 0.87 ± 0.04 (median, 0.88), mean HD = 0.98 ± 0.37 mm (median, 0.89 mm).23 Moreover, our proposed automatic method had higher accuracy for all total SC, WM, and GM compared with the iterative nonlocal STAPLE algorithm in our AMIRA images (all, P < .001).
Discussion
Visualization of the SC GM in MR imaging has been hampered by technical difficulties until recently.8 Despite technologic advancements, segmentation of SC compartments remains a challenge.24 In this work, we successfully deployed the novel MR imaging approach AMIRA and a fully automatic variational segmentation algorithm with a shape prior modified for 3D data with a slice similarity before demonstrating a fully automated approach for segmentation of SC, GM, and WM.
In contrast to brain MR imaging, the environment of the SC presents additional challenges for MR imaging methods and inherently for SC segmentation. The greatest challenges include magnetic field inhomogeneities across the SC, cord curvature, shape and size, contact of the SC and the osseous canal, osteophytes causing focal changes in CSF flow dynamics within the canal, motion artifacts, Gibbs artifacts, partial volume effects, and B1 inhomogeneity.25⇓–27 The AMIRA sequence13 used is based on a 2D approach that is, generally, less motion-sensitive than a 3D sequence. It uses a relatively short acquisition time of 51 seconds per slice, which leads to a reduction of motion artifacts and is especially suitable for disabled patients with limited ability to lie still (eg, due to spasticity). The AMIRA approach makes use of a balanced steady-state free precession readout, which is inherently of low-flow sensitivity or inherently flow compensated.13 The inversion recovery preparation is global and nonselective; hence, it does not pose an issue for CSF flow sensitivity either.
Furthermore, because the SC has a small cross-section of roughly 1.3 × 0.7 cm and our slices were located close to the isocenter, effects of B1 inhomogeneity do not play a significant role for the present AMIRA images. The even smaller size of the SC GM presents additional difficulties for MR imaging methods, requiring submillimeter in-plane resolutions, especially for morphometry. Visualization and segmentation of the SC GM and WM are hampered by the similar relaxation times of the 2 SC compartments, limiting the use of conventional SC MR imaging for that purpose. Finally, the complex butterfly shape of the SC GM makes the segmentation of the structure a rather difficult task for computer-based segmentation methods. The AMIRA approach was able to produce SC images with a high GM/WM contrast in all participants. This was achieved in clinically feasible acquisition times (10.2 minutes for a 48-mm cervical SC segment).
Of 864 slices, only 9 were excluded due to image artifacts, mainly resulting from magnetic field inhomogeneities produced by bone structures (clavicles, scapulae, humeri, ribs, and so forth) as well as due to aliasing and motion artifacts. Although, these artifacts occurred in a rather small percentage of the acquired images (1%), they should be taken into account in future applications of the AMIRA approach. A further argument in favor of the use of AMIRA for SC GM and WM quantification is that our pipeline was able to deliver not only higher accuracy measures compared with a previous study24 demonstrating results from various MR imaging sequences and segmentation algorithms, but also better accuracy performance of an established algorithm on AMIRA compared with T2* MR images (see also below). This result may be an indirect indication of the superior quality of AMIRA compared with other sequences used so far for spinal cord GM and WM quantification. Nevertheless, due to AMIRA having a nonisotropic resolution, our MR imaging acquisition may have been more prone to partial volume effects, despite our slices being angulated individually in an orthogonal way to the course of the SC.
The proposed automatic segmentation method showed excellent precision in terms of inter- and intrasession reproducibility and was superior to the manual segmentation performed by experienced raters for all SC metrics, as measured by both CV and DSC. Our automatic method was also superior in terms of the HD for total SC, though it did not differ with regard to SC WM and GM. At the same time, the accuracy of the automatic method was high for total SC, GM, and WM, as measured by both the DSC and HD. Comparing the present data with results of the SC GM segmentation challenge,24 we achieved a superior mean GM DSC of 0.86 versus 0.80 performed by the deepSeg (https://pypi.org/project/deepSeg/) algorithm in the SC GM segmentation challenge dataset. This achievement could be potentially explained by the high quality of the AMIRA images and/or the use of a multicenter dataset within the challenge with results from various MR imaging sequences and segmentation algorithms. Application of the previously published iterative nonlocal STAPLE algorithm on our AMIRA images showed higher accuracy than the original work of Asman et al23 (SC GM: median DSC of 0.82 versus 0.75, median HD of 1.04 versus 2.5 mm), which was performed on T2*-weighted 3D gradient-echo images. While our atlases were constructed from a pool of around 800 samples, Asman et al had around 2000 available slices. Thus, the better accuracy seen here can be explained by a possible higher image quality in AMIRA images compared with T2*-weighted 3D gradient-echo images; however, a direct comparison of MR images within the same subjects was not performed. The shallow architecture of the proposed algorithm with only a few parameters may make it less prone to overfitting to the training set compared with a state-of-the-art deep neural network. However, a direct comparison of our method with deepSeg28 was not possible in this study.
Precision and accuracy of our automatic method was decreased in the order of total SC→WM→GM. This decrease may be caused by the accordingly decreasing size of WM and GM compared with total SC because small differences may be translated into a larger variance. Moreover, the more complex geometry of the GM and WM compared with total SC may be more prone to misclassification errors. Finally, despite the good image quality, signal contrast was stronger for SC/CSF compared with GM/WM, which, in turn, could have partly contributed to differences in total SC and GM segmentation. Moreover, a slightly lower reproducibility and accuracy of our measurements in more caudally acquired slices could also be identified, which may reflect a decrease in contrast intensity and a “noise” increase in AMIRA images acquired closer to the lungs and surrounded by overall greater body mass (thorax, shoulders, and arms) compared with the more rostral cervical SC.
Our automatic method also showed significantly lower intrasession than intersession variability for all SC metrics. However, GM intra- and intersession CVs were similar, with mean values ranging between 4.10% and 4.77%. Accordingly, our method demonstrated similar mean intra- and intersession CVs between 2.54% and 2.95% for WM. We, therefore, conclude that patient repositioning only slightly influences GM and WM area measurements; this conclusion provides evidence for the suitability of our automatic segmentation method in longitudinal settings.
In our work, minimal contrast adjustment differences in our manual segmentation led to a marked decrease of reproducibility, especially in GM area quantifications, as shown in the manual intra- and interrater measurements (mean CV up to 19.18%). Because the proposed method is fully automatic and requires no user-software interaction, it is devoid of additional variation produced by intra- and interrater variability. Therefore, our method provides significant advantages in large datasets or multicenter studies and, as mentioned above, may also be valuable in the longitudinal evaluation of individual patients (eg, patients with MS).
Compared with the manual reference standard, the automatic method slightly overestimated total SC and WM area, while underestimating the GM area. This result might be due to different intensity-thresholding in the manual segmentation compared with the automatic method. The caudal GM area increase shown in Fig 3 can be explained by the increased volume of motor cells of the cervical SC enlargement in the GM ventral horns, which innervate the upper limb muscles.
Although a fully automatic segmentation was not feasible on 12% of acquired SC slices, a semiautomatic approach with manual total SC segmentation and fully automatic GM and WM segmentation could be performed on those slices. This approach also showed high-accuracy measurements with mean DSC of ≥0.83 in both GM and WM. However, compared with the fully automatic method on the initially nondiscarded slices, a slight accuracy decrease was observed, which could be interpreted in terms of a lower image quality of those AMIRA images. Nevertheless, these results demonstrate a relative robustness of our automatic approach even in MR images of suboptimal quality, which are a rather common phenomenon in clinical routine.
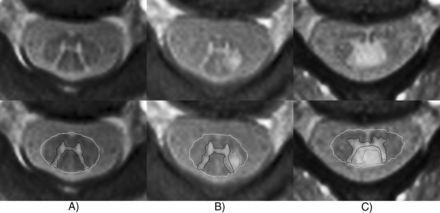
The present work focused on SC GM and WM segmentation using AMIRA images of healthy controls. Nevertheless, the motivation of our research is to deploy this method in patient data (eg, patients with MS) for the development of a potential widely applied MR imaging biomarker. Exemplary segmentation of data of patients with MS (not shown in detail here) showed that lesion appearance was similar to that of GM and therefore challenged the algorithm where lesions did not respect the GM boundaries (Fig 6). In future work, we intend to adjust the current method to address its current limitations. As an alternative approach, we plan to apply a deep learning–based segmentation approach on pathologic images as already performed on the data of healthy subjects.28,29
Examples of segmentations of representative patients with MS. The thick continuous line indicates automatic segmentation; the dashed line, manual reference standard. A, A 54-year-old female patient with MS. Rostral cervical SC slices of the C1/C2 level without focal lesions. Automatic segmentation highly corresponds to the manual reference standard. B, A 32-year-old male patient with MS. Rostral cervical SC slice of the C2 level with a focal posterolateral lesion fused with the left posterior gray matter horn. Automatic segmentation misclassifies the focal lesion as SC GM. C, A 33-year-old female patient with MS. A cervical SC slice of the C3/C4 level with a focal posterior lesion fusing with the posterior SC GM horns and the central SC GM commissure. Automatic segmentation misclassifies the focal lesion as SC GM and CSF.
Conclusions
The AMIRA sequence is presented as a time-efficient and reproducible MR imaging approach within the cervical cord. Our fully automatic segmentation method for SC GM and WM demonstrated further high reproducibility and accuracy. We were able to show that a shallow algorithm produces state-of-the-art GM-WM segmentation results on the AMIRA data. It is therefore suitable in large longitudinal studies investigating upper cervical SC volumes. Reproducibility measures of this work could be further used for effect size calculations of SC compartment metrics for studies using the same processing approach. In future work, we will address the use of deep learning approaches, as demonstrated in recent studies.28
Footnotes
C. Tsagkas and A. Horvath contributed equally as first authors.
Disclosures: Charidimos Tsagkas—RELATED: Grant: Swiss National Science Foundation (grant No. 320030_156860) and the Foundation for Sponsorship of Gastroenterological and General Clinical Research as well as of Medical Imaging (application ID 02/2015); UNRELATED: Grants/Grants Pending: University of Basel, Forschungsfonds für exzellente Nachwuchsforschende, pending. Antal Horvath—RELATED: Support for Travel to Meetings for the Study or Other Purposes: Swiss National Science Foundation.* Simon Pezold—UNRELATED: Grants/Grants Pending: Novartis Research Foundation.* Matthias Weigel—RELATED: Grant: Swiss National Science Foundation, Comments: grant No. 320030_156860, Method Development for MRI of Spinal Cord*; UNRELATED: Employment: Department of Biomedical Engineering, University of Basel, Switzerland, Comments: postdocterate; Patents (Planned, Pending or Issued): University Hospital Freiburg, Freiburg, Germany, Comments: patent for Optical Motion Correction, neither relevant nor used in the present work; Travel/Accommodations/Meeting Expenses Unrelated to Activities Listed: International Society for Magnetic Resonance in Medicine, European Society for Magnetic Resonance in Medicine and Biology, MR Compact, Comments: travel reimbursement and accommodation for invited, scientific talks about MRI physics. Ludwig Kappos—UNRELATED: Board Membership: Actelion, Almirall, Bayer AG, Biogen, Celgene/Receptos, df-mp, Excemed, Genzyme, Japan Tobacco, Merck, Minoryx, Mitsubishi Tanabe Pharma, Novartis, Roche, Sanofi-Aventis, Santhera, Teva Pharmaceutical Industries, Vianex*; Consultancy: Actelion, Almirall, Bayer AG, Biogen, Celgene/Receptos, df-mp, Excemed, Genzyme, Japan Tobacco, Merck, Minoryx, Mitsubishi Tanabe Pharma, Novartis, Roche, Sanofi-Aventis, Santhera, Teva Pharmaceutical Industries, Vianex*; Grants/Grants Pending: Bayer AG, Biogen, Novartis, Roche, the Swiss MS Society, the Swiss National Research Foundation*; Payment for Lectures Including Service on Speakers Bureaus: Allergan, Almirall, Baxalta, Bayer AG, Biogen, CSL Behring, Desitin, Excemed, Genzyme, Merck, Novartis, Pfizer, Roche, Sanofi-Aventis, Teva Pharmaceutical Industries*; Payment for Development of Educational Presentations: Allergan, Almirall, Baxalta, Bayer AG, Biogen, CSL Behring, Desitin, Excemed, Genzyme, Merck, Novartis, Pfizer, Roche, Sanofi-Aventis, Teva Pharmaceutical Industries.* Till Sprenger—RELATED: Grant: Swiss National Science Foundation*; UNRELATED: Consultancy: Actelion, ATI, Biogen, Desitin, electroCore, Sanofi Genzyme, Novartis, and Mitsubishi Tanabe Pharma Europe*; Grants/Grants Pending: EFIC-Grünenthal, Novartis Switzerland, the Swiss Multiple Sclerosis Society*; Payment for Lectures Including Service on Speakers Bureaus: Sanofi Genzyme, Biogen, Novartis, Teva Pharmaceutical Industries.* Oliver Bieri—RELATED: Grant: Swiss National Science Foundation*; UNRELATED: Grants/Grants Pending: Swiss National Science Foundation.* Katrin Parmar—RELATED: Grant: Swiss National Science Foundation*; UNRELATED: Grants/Grants Pending: Baasch Medicus Foundation; Payment for Lectures Including Service on Speakers Bureaus: Excemed, Novartis Switzerland*; Travel/Accommodations/Meeting Expenses Unrelated to Activities Listed: Novartis Switzerland. *Money paid to the institution.
This work was supported by the Swiss National Science Foundation (grant No. 320030_156860) and the Foundation for Sponsorship of Gastroenterological and General Clinical Research as well as of Medical Imaging (application ID 02/2015).
Indicates open access to non-subscribers at www.ajnr.org
REFERENCES
- Received November 26, 2018.
- Accepted after revision June 25, 2019.
- © 2019 by American Journal of Neuroradiology