Abstract
BACKGROUND AND PURPOSE: LINCL is a uniformly fatal lysosomal storage disease resulting from mutations in the CLN2 gene that encodes for tripeptidyl peptidase 1, a lysosomal enzyme necessary for the degradation of products of cellular metabolism. With the goal of developing quantitative noninvasive imaging biomarkers sensitive to disease progression, we evaluated a 5-component MR imaging metric and tested its correlation with a clinically derived disease-severity score.
MATERIALS AND METHODS: MR imaging parameters were measured across the brain, including quantitative measures of the ADC, FA, nuclear spin-spin relaxation times (T2), volume percentage of CSF (%CSF), and NAA/Cr ratios. Thirty MR imaging datasets were prospectively acquired from 23 subjects with LINCL (2.5–8.4 years of age; 8 male/15 female). Whole-brain histograms were created, and the mode and mean values of the histograms were used to characterize disease severity.
RESULTS: Correlation of single MR imaging parameters against the clinical disease-severity scale yielded linear regressions with R2 ranging from 0.25 to 0.70. Combinations of the 5 biomarkers were evaluated by using PCA. The best combination included ADC, %CSF, and NAA/Cr (R2 = 0.76, P < .001).
CONCLUSIONS: The multiparametric disease-severity score obtained from the combination of ADC, %CSF, and NAA/Cr whole-brain MR imaging techniques provided a robust measure of disease severity, which may be useful in clinical therapeutic trials of LINCL in which an objective assessment of therapeutic response is desired.
ABBREVIATIONS:
- FA
- fractional anisotropy
- GM
- gray matter
- LINCL
- late infantile neuronal ceroid lipofuscinosis
- MRIDSS
- MR imaging disease-severity score
- PCA
- principal component analysis
Lincl, a form of Batten disease, is a progressive uniformly fatal lysosomal storage disorder resulting from mutations in the CLN2 gene with predominantly neurologic symptoms.1 The CLN2 gene encodes for tripeptidyl peptidase 1 (TPP-I), a lysosomal protease.2 In affected children, undegraded products of cellular metabolism accumulate within lysosomes over several years, eventually causing cell death.3 Histologically, the lysosomes are characterized by the presence of autofluorescent ceroid lipofuscin. Neurologic symptoms of the disease begin to appear at 2–4 years of age and progressively worsen, leading to death by of 8–12 years of age. Diagnosis is traditionally made after the appearance of one of several clinical features, including retinopathy, motor abnormalities, epilepsy, or dementia.4 Definitive diagnosis of LINCL is made through enzymatic testing of skin biopsies or blood lymphocytes and mutation identification through molecular genetic testing.5,6
LINCL is currently incurable. The current standard of care is palliative in response to clinical symptoms, though research into novel therapeutic strategies, including gene transfer at our institution, is ongoing.6,7 Assessment of disease severity is made during subject examination via neurologic rating systems. One such scale developed at our institution assigns individual scores ranging from 0 to 3 to each of 4 neurologic functions, including feeding, motor, gait, and language development, with higher scores indicative of greater functionality in each area.8 The sum total of the 4 scores is used as a marker for overall disease severity, with the maximum score of 12 indicative of a healthy individual.
Previous MR imaging studies of LINCL displayed marked cortical atrophy and CNS volume loss with disease progression. Severe cerebellar atrophy, along with loss of cortical neurons, axons, and white matter myelin, was also documented.9 MR spectroscopy showed decreased NAA/Cr metabolite ratios and increased myo-inositol/creatine ratios compared with healthy controls.10 We have previously shown that increased ventricular volume and increased apparent whole-brain–water self-diffusion diffusion coefficients (ADC) are associated with a decreasing LINCL score.8,11
To develop more quantitative biomarkers of disease progression that could also be used to assess the efficacy of gene transfer for the CNS manifestations of LINCL, we directed the present study toward a comprehensive evaluation of a total of 5 quantitative MR imaging biomarkers. While information was available on a voxel-by-voxel basis for each of the methods, we evaluated an automated objective assessment of disease progression via analysis of whole-brain histograms. Each of the 5 quantitative MR imaging techniques interrogated different aspects of brain morphometry or metabolism and included the following: 1) the ADC of water, a measure of tissue integrity related to the restriction of the free molecular motion of water by cellular membranes; 2) diffusion FA, a measure of the anisotropic diffusion of water molecules, which is related to the degree of myelination in white matter independent of the ADC; 3) T2 relaxation times, an important basis for brain contrast in clinical MR imaging; 4) the volume percentage of CSF (%CSF), a measure of brain atrophy; and 5) NAA/Cr metabolite ratios, a marker for neuronal function.12⇓–14 Each of the 5 MR imaging biomarkers allows intersubject comparison or assessment of serial studies on a single subject. The goal of this study was to determine whether the combination of MR imaging–derived biomarkers was feasible to routinely acquire in a single scanning session in this patient population and whether it was relevant to calculating an overall disease-severity score.
Materials and Methods
Study Population
This study was conducted under a research protocol approved by the institutional review board at Weill Cornell Medical College. Parents or guardians signed informed consent. All subjects were diagnosed both by phenotypic findings and genetic analysis. Thirty MR imaging datasets were acquired from 23 subjects (2.5–8.4 years of age; median, 4.8 years; 8 male/15 female). Seven subjects were scanned at 2 time points as part of a separate therapeutic trial for LINCL but were untreated at the time of the scans. To participate in this study, the subjects' genotypes included at least 1 of the most common CLN2 mutant genes as outlined in On-line Table 1.15,16 The subjects' LINCL scores (0–12 scale) were 6.0 ± 2.5 on average and ranged from 1.5 to 11. All subjects were evaluated by using the Weill Cornell LINCL Disease Severity Scale as described above by 4 observers.8 Clinical evaluations were performed within 2 days of the corresponding imaging examinations.
Pearson correlation coefficients (R2) for individual MRI biomarkers (top row) versus the LINCL score and biomarker combinations performed using PCAa
MR Imaging Acquisition Techniques
All imaging data were acquired by using a 3T HDx MR imaging system (GE Healthcare, Milwaukee, Wisconsin) with an 8-channel head resonator. Standard-of-care clinical imaging was appended to the research study and included axial T1-weighted, T2 FLAIR, and coronal T2-weighted imaging series. The research sequences included single-shot axial diffusion-weighted echo-planar imaging for the ADC acquisition with b-values of 0 and 1000 s/mm2, a matrix size of 128 × 128, a 20.0-cm FOV, a 4.0-mm section thickness, and a 0.4-mm section gap. Diffusion tensor imaging for FA acquisition was performed by using an echo-planar pulse sequence with a b-value of 800 s/mm2 and 33 gradient directions. A matrix size of 128 × 128, a 25.6-cm FOV, and a 2.0-mm section thickness resulted in isotropic voxels with dimensions of 2.0 × 2.0 × 2.0 mm. For T2 mapping, a multiecho spin-echo sequence was used with TE of 20, 40, 60, and 80 ms. The pulse sequence TR was set at 1000 ms to limit the overall acquisition time of the series. Images at each TE were used for exponential fitting of the T2 value on a voxel-by-voxel basis. A total of 32 axial sections with a matrix size of 256 × 192, a 20.0-cm FOV, a section thickness of 4.0 mm, and a section gap of 0.4 mm were used to cover the whole brain. A sagittal high-resolution isotropic 3D BRAin VOlume imaging (BRAVO) sequence was applied with 1.0 × 1.0 × 1.0 mm spatial resolution for the calculation of %CSF volume. This sequence used a fast 3D-gradient echo technique with a TR of 12 ms, a TE of 5 ms, a TI of 450 ms, and an acceleration factor of 2.
Finally, for NAA/Cr measurements, proton spectroscopic imaging data were acquired with water suppression by using a spin-echo–based 4-section chemical shift imaging sequence and a TR of 2300 ms with a TE of 280 ms.17 A FOV of 20.0 cm and a chemical shift imaging matrix of 24 × 24 resulted in a voxel size of 0.83 × 0.83 × 1.5 cm (1.0 mL) in 16 minutes of scanning time. Pericranial lipid contamination was reduced by using octagonal outer volume suppression bands. All subjects were maintained under general anesthesia as a standard of care throughout the imaging procedures and were continuously monitored by an anesthesiologist. All of the above methods were applied to each subject in a total image examination time of approximately 75 minutes.
MR Imaging Analysis Techniques
Parametric images of the ADC were calculated by using processing software provided with the scanner. Fractional anisotropy values were calculated for each voxel also by using vendor-supplied software. Image segmentation into GM, white matter, and CSF components was performed by using the fMRI of the Brain Software Library (http://www.fmrib.ox.ac.uk/fsl/).18,19 The Brain-Extraction Tool was used for skull-stripping followed by segmentation by using FMRIB's Automated Segmentation Tool with 8 iterative passes for bias field correction. Coregistration of the FA and T2 maps with the segmentation masks was achieved by aligning the FA b=0 s/mm2 and T2 TE = 20 ms images with the 3D BRAVO scans, respectively. Spectroscopic image acquisition used N-acetylaspartate at 2.02 ppm as a reference peak. A susceptibility correction used a point-by-point cross-correlation between subsequent spectra and the reference spectrum for alignment. Peak areas were integrated by using the XSOS spectroscopic analysis software package developed in-house (D.C.S., X.M.), and the NAA/Cr ratio was calculated for all voxels.
An image mask based on a low-signal-intensity threshold was used for ADC measurements to create histograms containing all voxels within the brain while discriminating against regions containing only noise. The contribution from CSF to the ADC maps was prevented because the CSF and brain parenchyma compartments were easy to discern in the histograms as we described previously.11
The mode of the parenchymal peak in the histogram was used as the relevant variable for the ADC histogram. To reduce the CSF contamination from the FA and T2 histograms, we isolated contributions from gray and white matter. White matter FA histograms were produced by taking the product of the binary white matter segmentation mask and FA maps. Gray matter T2 maps were calculated from voxels identified via the product of the gray matter segmentation mask and T2-weighted images. Application of the white and gray matter masks to the FA and T2 images greatly reduced CSF contamination in these measures, allowing the mean of the histogram to be used for characterization. For spectroscopy, all voxels in the brain excluding those in the lateral ventricles were combined to produce a histogram of NAA/Cr values.
Statistical Methods
PCA was performed on the MR imaging dataset by using Matlab (R2011a; MathWorks, Natick, Massachusetts). PCA determines the direction of greatest variability of the data in the MR imaging biomarker variable space using a linear combination of the biomarkers. The data for each component were first standardized by subtracting the mean and dividing by the SD, thereby allowing combination of biomarkers with different units. Next, the singular value decomposition method was used to solve for the principal components. All combinations of n biomarkers were tested with n ≤ 5. The output of the analysis was an n-dimensional-unit vector of coefficients for the direction of greatest variability (PC1), followed by an n-dimensional-unit vector designating the direction of next greatest variability orthogonal to PC1 (PC2), and so on until a complete set of n-orthogonal-basis vectors was specified. We then defined an MR imaging–based disease-severity score to be equal to the linear combination of the n-biomarkers weighted by the PC1 coefficients. This scoring system had a scale that was determined by minimizing the sum of squared differences with the clinical LINCL scores and thus was similar to the 0–12 range of the clinical LINCL scale. Finally, we determined the linear correlation of this score with the clinical LINCL score by calculating Pearson correlation coefficients (R2).
Results
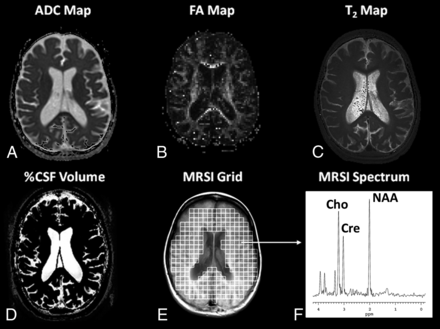
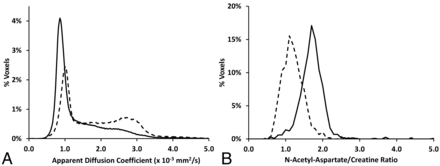
Representative images are shown in Fig 1 for a subject (BD-04) at 5.2 years of age with a clinical LINCL score equal to 3.0, showing enlarged ventricles. In general, the image data were of high quality and specifically free of major motion artifacts, owing to the general anesthesia that was administered throughout the study. In Fig 2, examples are shown of histograms from the ADC and NAA/Cr biomarkers for subjects with LINCL scores of 3 and 9 exhibiting marked differences due to disease progression.
Representative parametric images from each of the MR imaging biomarkers for subject BD-04 at 5.2 years of age with a clinical LINCL score of 3.0. A, ADC. B, FA. C, T2 relaxation time. D, CSF segmentation for %CSF volume calculation. E and F, Spectroscopic imaging grid and representative spectrum. Peak assignments are Cho, Cr, and NAA.
Whole-brain histograms comparing a subject in the early stages of LINCL (BD-02, clinical LINCL score = 9, solid lines) with one with more advanced disease (BD-09, clinical LINCL score = 3, dotted lines). A, ADC; note the bimodal distribution of the LINCL = 3 subject, indicative of increased ventricular volume and tissue atrophy; the parenchymal peak is on the left. B, N-acetylaspartate-to-creatine ratio.
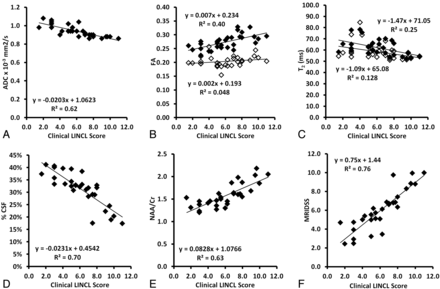
In general, the ADC (R2 = 0.62, P < .001), gray matter T2 relaxation time (R2 = 0.25, P < .005), and %CSF volume (R2 = 0.70, P < .001) all increased with disease progression, while the white matter FA (R2 = 0.40, P < .001) and NAA/Cr (R2 = 0.63, P < .001) decreased with increasing disease severity. Most surprising, FA and T2 were more weakly correlated with LINCL disease severity than the other components, even when segmented into gray and white matter contributions (Fig 3A-E). Indeed, we found the highest correlation between the MR imaging–based score and clinical disease severity when combining only 3 components: ADC, %CSF, and NAA/Cr with R2 = 0.76, P < .001 (Table). Therefore we did not include FA or T2 in our final disease-severity score (Fig 3F).
The results of histogram analysis are shown for all MR imaging biomarkers and all subjects including the ADC (A); white matter (solid diamonds, B) and gray matter (hollow diamonds) FA; gray matter (solid diamonds) and white matter (hollow diamonds) T2 relaxation time (C); %CSF (D); NAA/Cr ratio (E); and MRIDSS calculated from the combination of ADC, %CSF, and NAA/Cr, each plotted versus the clinical LINCL score (F).
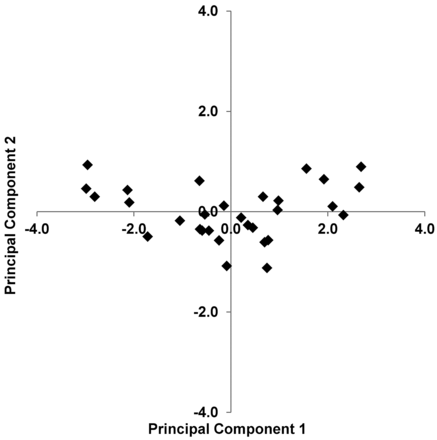
A plot of PC1 versus PC2 for all 30 subjects is shown in Fig 4. For the ADC, %CSF, and NAA/Cr combination, PC1 accounted for 86.0% of the variability in the data, with PC2 accounting for an additional 9.8%. The vector of coefficients for PC1 with n = 30 subjects, normalized to unit length, was equal to 0.56, 0.59, −0.58 and was used to define the MRIDSS as
 with the mean and SD of each biomarker given by ADC (0.94 ± 0.06 × 10−3 mm2/s), %CSF (31.4% ± 6.8%), and NAA/Cr (1.58 ± 0.26). The factor −1.0 was inserted in the first equation so that the MRIDSS and clinical LINCL score both assigned higher scores to healthier subjects. The additive factor B was chosen so that the MRIDSS had the same mean value as a parent population of clinical LINCL scores, that is, B = 6. Next, note that application of the factor A is equivalent to linearly scaling the unit vector of coefficients of PC1. Because we chose a scaling factor such that A minimized the square of the differences between the MRIDSS and the clinical LINCL scores, we found A = 1.33. Equation 1 with the above coefficients is plotted in Fig 3F, and all numeric data for individual subjects is provided in On-line Table 1. The predictive accuracy of the MRIDSS as estimated by the root mean squared predictive error was 1.49, based on the SD of the residuals of equation 1 with the clinical LINCL score.
with the mean and SD of each biomarker given by ADC (0.94 ± 0.06 × 10−3 mm2/s), %CSF (31.4% ± 6.8%), and NAA/Cr (1.58 ± 0.26). The factor −1.0 was inserted in the first equation so that the MRIDSS and clinical LINCL score both assigned higher scores to healthier subjects. The additive factor B was chosen so that the MRIDSS had the same mean value as a parent population of clinical LINCL scores, that is, B = 6. Next, note that application of the factor A is equivalent to linearly scaling the unit vector of coefficients of PC1. Because we chose a scaling factor such that A minimized the square of the differences between the MRIDSS and the clinical LINCL scores, we found A = 1.33. Equation 1 with the above coefficients is plotted in Fig 3F, and all numeric data for individual subjects is provided in On-line Table 1. The predictive accuracy of the MRIDSS as estimated by the root mean squared predictive error was 1.49, based on the SD of the residuals of equation 1 with the clinical LINCL score.
PC1 versus PC2 for all n = 30 data points for the combination of ADC, %CSF, and NAA/Cr. The numeric values for PC1 and PC2 are provided in On-line Table 1.
Discussion
Routine T1- and T2-weighted MR imaging pulse sequences are frequently insensitive to pathophysiologic changes in neurodegenerative diseases.20 Conversely, several studies using quantitative measurements of each of the 5 MR imaging biomarkers chosen for this work have shown independent utility in assessing underlying physiologic parameters in these diseases.11⇓⇓–14,21,22
Multiparametric MR imaging has been applied to various disease states, primarily focusing on oncologic applications.23⇓–25 Validation of multiparametric datasets has typically relied on binary categorization of histologic tissue type as containing either benign or malignant cells. Relying on this dichotomization of tissue, receiver operating characteristic curves and specific cutoff thresholds are typically defined for each MR imaging biomarker to calculate specificity or sensitivity, depending on the disease state. Application of multiparametric MR imaging has been limited in assessing continuous progression of severity such as seen in neurodegenerative diseases.
The histogram analysis methods described in the present study allowed us to sidestep an important problem in handling multiparametric imaging datasets, namely how to compare results from different data-acquisition series that are optimally acquired with a range of spatial resolution or section orientations. In our previous work, by using the ADC as a measure of disease severity, we developed a detailed analysis of multiple histogram peaks, as are evident in Fig 2A, that included consideration of the important problem of partial volume averaging in imaging.11,26 For the present work, we simplified the analysis, calculating only the modes and means of histograms, with the aim of enhancing the utility of the methods in clinical trials. Specifically, the image analysis is automated and is, for example, independent of user-specified region-of-interest analysis. Characterization of histograms without advanced modeling tools or sophisticated user intervention should facilitate broader use of the technique.
Each of the 5 MR imaging biomarkers in this study contains complementary information. A significant correlation of each MR imaging biomarker was found with clinical LINCL scores, with P < .005 in all cases. The imaging metric obtained from the combination of whole-brain ADC, %CSF, and NAA/Cr variables resulted in a generalized measure of disease severity and offers a time-savings of approximately 20 minutes in image acquisition relative to inclusion of all 5 biomarkers. The metric is a noninvasive objective measure requiring no introduced MR imaging contrast agents. It may prove useful in future serial assessments of subjects undergoing treatment.
Note that since the clinical score is subjective and not a definitive standard, for example, as one might obtain from histology, it is unclear that a given discrepancy in the correlation is necessarily attributable to deficiency in the combined MR imaging score or any of the individual components. In an attempt to minimize clinical variability, we had multiple experienced examiners evaluate the LINCL score for each patient and then average their scores. Aside from being an objective measure of disease severity, a key advantage of our MR imaging method is the potential ability to refine the evaluation to specific brain areas in ways that are beyond the capability of clinical examinations. Also, the fact that 3 independent biomarkers were used in combination is likely to make the MRIDSS more robust with respect to variability due to different scanner platforms.
With regard to the comparison of the calculated regression lines for the MR imaging variables in subjects with LINCL relative to the trends in age-matched normative data, there are significant differences according to the literature.27⇓⇓–30 Normative measures of ADC showed a decrease with pediatric age as opposed to that seen with progression of LINCL.27 Likewise, WM FA increased with normal aging in the pediatric population as opposed to decreases in WM FA seen with LINCL disease progression.27 Whole-brain T2 histograms decreased with age in pediatric populations as opposed to the increase in gray matter T2 values found with advanced LINCL.28 NAA/Cr ratios did not change significantly with age during early childhood and adolescence, while significant decreases were found with advancing LINCL severity.29 CSF volume remained constant at values ranging from 7% to 9% from early childhood to adolescence in healthy subjects compared with 3–4-fold increases in the %CSF seen in advanced LINCL.30
Finally, the methods developed for this work are, of course, quite general and, specifically, are not limited to any single brain pathology. The measurement of multiple independent biomarkers via noncontrast MR imaging in an acceptable scanning time of ≤75 minutes, combined with a simple automated histogram and principal component analysis, may provide a robust methodology for assessment of disease severity in multicenter clinical therapeutic trials of a variety of brain diseases.
Conclusions
A quantitative noninvasive MR imaging–based disease severity score for late infantile neuronal ceroid lipofuscinosis has been presented. The metric combines data from brain-water apparent diffusion coefficients, the volume percentage of CSF, and N-acetylaspartate-to-creatine metabolite ratios. The methods used can be adapted to run on multiple scanner platforms in a straightforward manner.
Footnotes
Disclosures: Stephen M. Kaminsky—RELATED: Grant: NIH.* Barry Kosofsky—RELATED: Grant: NINDS.* Kyle D. Rudser—RELATED: Grant: NIH, Comments: partial support from NIH grant U54NS065768.* Dikoma C. Shungu—UNRELATED: Consultancy: Roche Product Limited, Florida Hospital. Stefan Worgall—RELATED: Grant: NIH.* Kaleb Yohay—RELATED: Grant: NINDS.* *Money paid to the institution.
This work was supported in part by grants from the National Institutes of Health (R01NS061848, UL1RR024996, and U54NS065768) and from the Partnership for Cures, Chicago, Illinois.
Indicates open access to non-subscribers at www.ajnr.org
References
- Received April 19, 2012.
- Accepted after revision June 14, 2012.
- © 2013 by American Journal of Neuroradiology