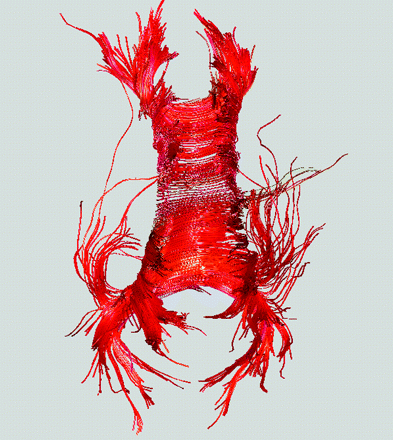
Abstract
SUMMARY: There have been numerous reports documenting the graphic reconstruction of 3D white matter architecture in the human brain by means of diffusion tensor MR tractography. Different from other reviews addressing the physics and clinical applications of DTI, this article reviews the computational principles of tractography algorithms appearing in the literature. The simplest voxel-based method and 2 widely used subvoxel approaches are illustrated first, together with brief notes on parameter selection and the restrictions arising from the distinct attributes of tract estimations. Subsequently, some advanced techniques attempting to offer improvement in various aspects are briefly introduced, including the increasingly popular research tracking tool using HARDI. The article explains the inherent technical limitations in most of the algorithms reported to date and concludes by providing a reference guideline for formulating routine applications of this important tool to clinical neuroradiology in an objective and reproducible manner.
Abbreviations
- CC
- corpus callosum
- DTI
- diffusion tensor imaging
- FA
- fractional anisotropy
- HARDI
- high angular resolution diffusion imaging
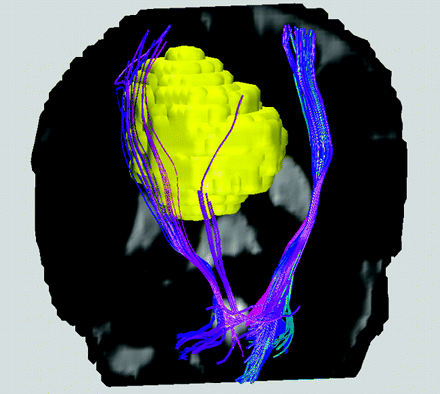
At the beginning of the 21st century, the radiologic community witnessed the tremendous technical progress toward noninvasive investigations of the white matter architecture in the central nervous system. Thanks to the advancement of DTI plus the ever-growing computer technology of the so-called tractography methods, a comprehensive visualization of the white matter fiber bundles and their relationships to tumors (Fig 1) can be readily displayed without the need for any surgery.1 Needless to say, the potential implications in presurgical planning are huge, particularly if the related technology can be executed objectively and automatically on a daily basis, with the reliability approaching a level that is highly acceptable in routine practice.
Colored white matter tracts rendered for a patient with a falx meningioma (yellow) on the right hemisphere near the location of the precentral primary motor cortex, superimposed on a coronal T2-weighted image. Blue is used for the corticospinal tracts, for which the right-sided fiber bundles are displaced by the presence of the meningioma. Note that though differential diagnosis for meningioma is relatively straightforward using conventional T1- and T2-weighted MR imaging, hence making diffusion MR tractography unnecessary, the example shown here at least demonstrates the validity of the displaced tracts reconstructed from DTI data.
This article attempts to provide a pedagogic overview of the aspects of diffusion MR tractography that clinical neuroradiologists may find helpful for routine use. Because an overview of data acquisition via DTI and potential clinical applications has been provided in several review articles,1–3 only the computational means to perform the tractography will be introduced here. In addition, only selective methods will be mentioned, because detailed comprehensive explanations about the massive amount of newly proposed algorithms are neither necessary nor insightful. At the same time, the ultimate restrictions originating from the computation methods will be addressed. With the basic concepts understood, extensions to more sophisticated algorithms would ideally be easily comprehensible. Finally, we conclude with a suggestive guideline for optimal use of diffusion MR tractography in clinical neuroradiology, emphasizing particularly the preparatory work that ought to be done before routine application. The mathematics in this entire article, though inevitable, will be kept at a minimum to ensure readability.
Diffusion Tensor Ellipsoid
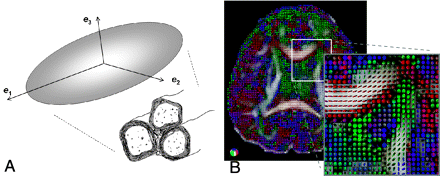
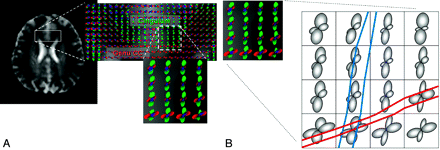
Because the primary focus of this article is on the principles and limitations of tractographic computations, review of physical principles related to DTI by using echo-planar imaging will be omitted. Interested readers are referred to the series of 2 review articles by Mukherjee et al2,3 for a comprehensive overview of the physical principles and scanning optimization for DTI. At this point, it suffices to simply remember that the diffusion tensor ellipsoid reflects the tendency of diffusion along various directions and that this information can be obtained from the original diffusion-weighted images on a voxel-by-voxel basis (Fig 2). Therefore, the direction of the longest axis of the ellipsoid, mathematically derived as the principal eigenvector of the 3 × 3 tensor matrix, corresponds to the major orientation of the axonal fiber bundle in the image voxel. The FA, a quantitative index expressed as a numeric value between 0 and 1, reflects the directionality (or equivalently, how elongated the ellipsoid is) of the tissue in the image voxel. Note that for most of the current tractography algorithms reported to date, the orientation and FA of the diffusion tensor constitute the sole information used to reconstruct the 3D white matter fiber tracts. Naturally, as will become clear in the following sections, the ultimate limitation of tract estimations essentially originates from the inadequacy of the information provided by tensor orientation and FA.
A, Schematic drawing showing the relationship between the diffusion tensor ellipsoid and the underlying white matter fiber bundle. The direction of the longest axis of the ellipsoid, mathematically called the principal eigenvector of the diffusion tensor (e1), corresponds to the major fiber direction where water molecules exhibit the fastest diffusion due to lack of a barrier. This direction is the most important information used in all tractography algorithms, whereas the other 2 axes (e2 and e3) of the diffusion tensor ellipsoid are used in only some of the methods. B, The diffusion tensor ellipsoid can be obtained on a voxel-by-voxel basis. The color-coding scheme is shown on the lower left corner.
Basic Principles of Tractography
Voxel Connection Based on Major Directions
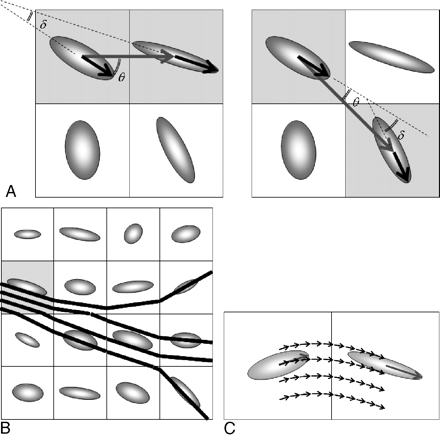
One of the simplest methods for performing tractography is to connect the neighboring image voxels that are thought to belong to the same white matter fiber tract, starting from 1 preassigned voxel called the “seed” voxel. This can be achieved by examining the directional consistency of 2 relationships: one between the principal eigenvectors of the 2 neighboring voxels under examination for connectivity and the other between the fiber direction and the vector connecting the 2 voxels (Fig 3A).4 The angle θ between 2 vectors ā and b̄ can be easily calculated with the following inner product formula:
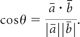
A, Connection of fiber tracts at the voxel level can be achieved by examining the directional consistency of 2 relationships: one between the principal eigenvectors (thick black arrows, angle δ) of the 2 voxels under examination for connectivity (shaded) and the other between the fiber direction and the vector connecting the 2 voxels (thick gray arrow, angle θ). If either of the 2 angles is larger than a prespecified threshold (eg, 18°), as shown on the left, these 2 voxels are not considered connected. When both of these angles are smaller than the threshold, connections are formed as on the right. The examination process then proceeds to the next voxel to continue the tracking. B, Schematic drawing illustrating a popular subvoxel tractography algorithm on a region consisting of 4 × 4 voxels. With seed points chosen in certain voxels (shaded), the tracts simply follow the direction of the principal eigenvector until reaching the voxel boundary, after which the tracts enter a neighboring voxel to continue the tracking process. Note that different seed points, even if placed in the same starting voxel, could lead to distinct tracking results as shown in this example. C, The computed tracts in subvoxel tractography can be made smoother by using smaller steps (one-tenth of the voxel width in this example) during the fiber tracking process. Successive alteration of the tract direction is performed (short black arrows) by using distance-weighted interpolation of the 2 principal eigenvectors (long gray arrows) of the diffusion tensor ellipsoids in the 2 neighboring voxels.
When both vectors are normalized to unit length (ie, length=1), the denominator vanishes and the examination for connectivity becomes computationally very economic. After examining the angular relationships between 1 certain voxel and all its neighboring voxels (totally 3 × 3 × 3 3D voxels), one forms connections by choosing those with angles smaller than a prespecified threshold. The process then proceeds to the next voxel to extend the fiber tracts until certain stop criteria are reached, followed by volume- or surface-rendering for 3D stereo visualization of the tracts. Two stop criteria most frequently used in current tractography algorithms are the turning angle and the FA value. Namely, for an acute turning white matter tract like the Meyer loop, the connection of 2 neighboring voxels would unlikely be formed at such a large turning angle. On the other hand, when the FA value of a voxel is less than a prespecified threshold, tract tracing should stop because the voxel under examination is likely to contain mostly gray matter or the CSF.
The definition of the voxel neighborhood for the examination of connectivity can be extended from the simplest case of 3 × 3 × 3 to, for instance, 5 × 5 × 5. This extension would allow a “jump” of the reconstructed tract in case an underlying voxel contains an erroneously estimated fiber direction due to fiber crossing or noise contamination, at the expense of computational complexity. The optimal choice of the voxel neighborhood definition is certainly a function of the spatial resolution and the width of the fiber tracts under consideration.
Several issues are noted here regarding the so-called discrete tractography algorithm illustrated above, with similar considerations largely held true for many (though not all) other tractography algorithms.5 First, image acquisition with an isotropic voxel dimension is preferable for fiber tractography, though not absolutely necessary.6 For imaging data whose section thickness substantially exceeds the in-plane voxel width, the examination of the relative orientation of the vector connecting the 2 neighboring voxels, for example, would need separate calculations along the section direction. Second, determination of the voxel connection based only on the principal eigenvector of the diffusion tensor essentially means that the information regarding the second and the third eigenvectors is entirely excluded (except partially taken in the FA value used as one of the stop criteria). Possible usage of the entire tensor information in tract tracing will be explained in a later section. Third, the examination criteria of directional consistency for connecting voxels is in favor of reconstructing white matter tracts that do not exhibit sharp turning angles. Tracts known to show prominent directional turning, such as the Meyer loop, thus represent difficult cases for computational tractography algorithms.
Placement of the seed voxels is another factor that could influence the tractography results. To identify white matter tracts passing through certain regions of interest, one can preassign seed voxels in, for example, the internal capsule to start the tract tracing. Alternatively, the seed voxels can be placed globally within the entire brain region, such that all possible tracts within the range of computational considerations are detected at the expense of huge loads to the computations (hence called the “brute force” approach).7 Subsequently, tracts passing through the designated regions of interest are then picked up by filtering out those undesired ones.
Fiber Connection in the Subvoxel Coordinate System
Although conceptually straightforward, the simplified methods mentioned in the previous section are rarely used in practice for one obvious reason: The white matter tracts obtained therein are restricted to the original spatial resolution of the image acquisition. This is especially important because the signal-intensity-demanding echo-planar diffusion imaging sequence provides voxel widths of approximately 2 mm for the usual 128 × 128 acquisition matrix.3 Limited spatial resolution not only leads to loss of the smaller fiber bundles but also causes voxel-quantization errors in the tract orientation.2 Therefore, most of the tractography algorithms proposed in the literature actually attempt to trace fiber tracts in a subvoxel manner.
One of the first and rather popular methods uses subvoxel fiber tracking with the principle shown in Fig 3B.8,9 Starting with seed points placed at certain locations within a voxel, the tracts simply follow the principal eigenvector of the diffusion tensor ellipsoid until reaching the voxel boundary, after which the tracts enter a neighboring voxel to continue the tracking process. Stop criteria using the turning angle and FA value thresholds, just as in discrete tractography, are used to determine the termination of fiber tracking. Note that different seed points, even if placed in the same starting voxel, likely lead to distinct tracking results (Fig 3B). This approach, therefore, allows identification of numerous thinner fiber bundles exceeding the spatial resolution of the original images. During the tracking process, the computer software needs to record all the location coordinates of the tracking steps for every single tract, instead of simply highlighting certain voxels as in the discrete tractography approach. Consequently, memory requirements are much more demanding in these so-called continuous tractography algorithms.8
The white matter tracts can be made even smoother by using smaller steps during the tracking process. This can be achieved by assigning a predetermined step size (eg, one-tenth of the voxel width) and performing continual alterations of the tract direction according to the expected fiber orientation at each subvoxel location (Fig 3C). The fiber orientation at each subvoxel location, in turn, would need to be estimated by using the tensor field originally acquired at a much coarser spatial resolution. Distance-weighted interpolation is frequently used for this purpose: Namely, the fiber orientation should be closer to that of the principal eigenvector of the voxel when the voxel is close to the subvoxel location under consideration and it should be affected by other nearby principal eigenvectors in a decreasing manner as those voxels move farther away.8 One simple interpolation example may look like the following equation:
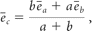 where ēc, ēa, and ēb are the vectors representing the fiber direction to be estimated and the principal eigenvectors of the 2 diffusion tensors used for the interpolation calculation, and a and b are the distances from the location to the centers of the 2 nearest voxels, respectively. A more sophisticated interpolation algorithm to minimize angular error accumulation can certainly be used.10 The equation above can also be extended to >2 voxels for a maximal inclusion of all the major fiber information available in the neighborhood.
where ēc, ēa, and ēb are the vectors representing the fiber direction to be estimated and the principal eigenvectors of the 2 diffusion tensors used for the interpolation calculation, and a and b are the distances from the location to the centers of the 2 nearest voxels, respectively. A more sophisticated interpolation algorithm to minimize angular error accumulation can certainly be used.10 The equation above can also be extended to >2 voxels for a maximal inclusion of all the major fiber information available in the neighborhood.
When stepping through a curve in the tensor field, the radius of curvature of the identified tract is strongly dependent on the step size (Fig 3C), one of the many tunable parameters in most of the current tractography algorithms. Obviously, a smaller step size allows the tractography program to follow more closely the curvature of the tracts, at the expense of increased computational loads. In addition, just as explained previously for discrete tractography, placement of the seed points to start the tracking process also has critical determining effects in subvoxel continuous tractography. Because there are virtually infinite possibilities in choosing the number, locations, and separation of the seed points, the resulting white matter tracts derived from the calculations may exhibit a somewhat different appearance, even if the same algorithms are used with identical choice of the region of interest to place the seed points. Regions showing branching or even diverging fibers are examples most likely demonstrating discrepancies in tractography results due to varieties in seed point assignments (Fig 3B).9,10
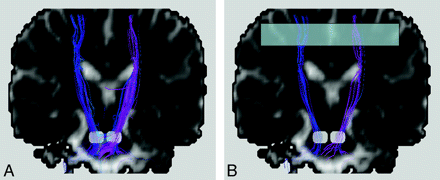
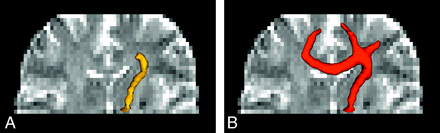
One method to increase the tract consistency is to select multiple regions of interest to retain the specific tracts that pass through all these regions. For instance, the white matter tracts within the corticospinal tracts responsible for motor function of the lower limbs can be isolated by encircling the medial precentral gyrus, the posterior limb of the internal capsule, and the cerebral peduncle in the midbrain. In this manner, unrelated tracts are filtered out, and the chance for false inclusion of spurious tracts is reduced. This technique is sometimes called the tract editing or virtual parcellation9,11,12 and is very useful in removing false-positive tracts (Fig 4). Certainly, the selection of regions of interest is based on prior knowledge of the neural transmission pathway. Application suitability in pathologic situations should, therefore, warrant careful attention.
Bilateral corticospinal tracts of a healthy subject with the 2 white regions on the coronal views selected to place the seed points. The obvious spurious tracts shown in A can be removed by selecting one more region of interest (the translucent box in B) to filter out unwanted tracts that do not pass through this region. This procedure is called tract editing and can be used in virtually all tractography algorithms. For this example, fiber assignment by continuous tracking is used.
One should keep in mind that tract editing is not only unable to reduce the false-negative rate but may also reduce the true-positive rate in tractography. Together with the dependency of tract curvature on step size as mentioned before, possible inability to follow a curve accurately could lead to a loss of tracts (ie, decreased sensitivity and underestimation of tract extent). The phenomenon is especially prominent for peripheral fibers that stray out of the main tract and later get filtered out by, for example, inappropriate tract editing. For neurosurgeons, this could be an important issue.
Adjustment of Tract Orientation by Using the Entire Tensor Information
Tract connection based solely on the principal eigenvectors of the diffusion tensors (ie, the longest axis of the ellipsoid) in the voxels ignores information contained in the second and third eigenvectors (ie, the short axes of the ellipsoid). In cases of disk-shaped ellipsoids whose 2 longer axes are roughly equal, as in the case of 2 intersecting fiber bundles roughly perpendicular to each other, the direction determination of the principal eigenvector is prone to prominent errors in the presence of noise. One method uses the diffusion tensor matrix multiplication to adjust for fiber direction.13 For a preassigned step size, the fiber orientation incoming vector, vin, encountering a voxel with diffusion tensor D is adjusted to produce the outgoing vector, vout, for the next tracking step, by using the following formula:
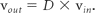
The physical meaning of the matrix multiplication can be viewed as a decomposition of the incoming vector into the coordinate system formed by the 3 eigenvectors of the diffusion tensor ellipsoid, with the 3 components weighted by the corresponding eigenvalues:
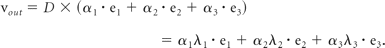
Consequently, after matrix multiplication, the tracking direction is altered somewhat toward the direction of the eigenvector with the largest eigenvalue. When the 2 larger eigenvalues are close in magnitude, the amount of deflection toward the 2 directions is nearly equal. In the special case of fiber-crossing that yields a nearly isotropic diffusion tensor (ie, FA ∼ 0), the equal weighting on the 3 orthogonal directions simply causes little change in the tracking direction. One may even use adaptive stepping to alter the step size according to local tensor characteristics for performance optimization.14
Certainly, deflection of the fiber tracking direction by using the entire diffusion tensor information can be extended to more sophisticated functions rather than the simple matrix multiplication, because over-reliance on the entire diffusion tensor is perhaps as unnecessary as reliance solely on the principal eigenvector for tractography. After all, anisotropy per se is only an index instead of the absolute measure of the directionality of the voxel under examination. Fiber bundles known to be highly unidirectional in the corpus callosum of healthy subjects, for instance, can exhibit regionally dependent FA values that may vary from 0.35 to 0.65 due to difference in the microarchitecture.15,16 As a consequence, nonlinear operations for tensor-based tractography, such as a reinforcement of alignment in the presence of a highly unidirectional tensor and a direct passing of the tracking vector when encountering an isotropic tensor, can be performed by examining the magnitudes of a series of shape-describing parameters of the diffusion tensor (called the linear, planar, and spheric coefficients) to select the best suitable operations for the voxel under examination.17 Interpolation of the tensor at subvoxel locations can also be used for refinements.10,17
More Advanced Tractography Algorithms
The number of tractography algorithms available for diffusion MR imaging keeps increasing at an incredible speed. Compared with those illustrated in the previous sections that use relatively straightforward computational approaches, sophisticated algorithms proposed at later times all claimed superiority in various aspects under certain conditions. While it is unnecessary to explain the complicated principles used in all the methods, it would be worthwhile to give a brief introduction to some of them to realize the potential advantages brought about by these means.
Fast-Marching Algorithms
“Fast-marching” refers to the computational process where the passage from 1 vector to another is done in a way similar to the propagation of a water wave from the seed point.5,18 The speed of the “wavefront” toward all directions is determined by the colinearity of the principal eigenvectors in the neighboring voxels, following which the direction of fiber tracking in a region-growing manner is assigned as the one with maximum propagation speed.19 In more advanced fast-marching algorithms, the propagation speed can also be modified on the basis of the magnitude of the eigenvalues rather than on the directions of eigenvectors alone.20 Other variations capable of resolving crossing fibers have also been proposed.21
Fiber Tracking via the Fluid Mechanics Analogy
The analogy of fiber tracking to the stream of viscous fluid flow inspired the use of computational fluid mechanics in tractography. The information from the diffusion tensor is included by a modeling process by using the Navier-Stokes equation, an equation well-known in fluid mechanics governing a wide range of physical phenomena, to simulate an artificial fluid flow through the entire imaging volume.22 Subsequently, the most likely connection paths between preassigned points in the imaging volume are estimated on the basis of certain metrics (the gradient of vector flow) derived from the fluid velocity vector field. Because the diffusion tensor anisotropy is inherently incorporated into the local “viscosity” of the fluid, there is no need for a white matter mask as used in many tractography algorithms.22
Global Optimization Approaches
Tracking of the white matter fibers by honestly following the principal eigenvectors of the diffusion tensors is prone to error accumulation when the estimation of direction is uncertain in the presence of noise.10 Consequently, as the distance from the seed point becomes larger, fiber tracking accuracy would decrease. Global optimization approaches were proposed to remedy this problem.23–25 In the simplest sense, the principles are analogous to curve fitting as opposed to connecting all points in a scatter plot. Often some modeling is performed, first for a predetermined geometric smoothness of the fiber tracts; then the shortest or the most suitable path is searched so that a general agreement with the principal eigenvectors can be reached.23 To facilitate the fitting calculation, one must define some “energy” and “penalty” and evaluate them iteratively to reach the balance between tract smoothness and tensor consistency.24 Needless to say, a huge number of tractography algorithms belong to this broad family.26 In general, better immunity to cumulative effects of noise can be expected.25
Tractography Incorporating A Priori Anatomic Knowledge
The fiber tracking algorithms mentioned in the previous sections are based solely on directional consistency or tract smoothness. In other words, judgment of fiber connection is computational rather than anatomic. Incorporation of prior anatomic information can be done via a multiple-step approach, where the application of the tractography computation is preceded by a global searching procedure to find the anatomic connections used to guide the subsequent fiber tracking process.27 With anatomic guidance, improvements in tracking accuracy are reported, especially in the region of complex fiber crossing. On the other hand, because normal anatomy is not to be extrapolated to pathologic situations, some studies even used anatomic templates constructed from a specific group of patients to assist tractography.28
Probabilistic Tractography
Algorithms that construct a 3D view of the white matter connection pathways are sometimes referred to as “deterministic tractography.” Rather than giving direct visualization of the fiber bundles, probabilistic tractography informs the users about the estimated probability distribution of how likely 2 or more certain regions are interconnected (ie, confidence mapping). The typical method to estimate the probability of fiber connection is to perform repeated image acquisitions from the same subject and, subsequently, use 1 specific algorithm on these datasets to obtain several tractograms.29 The probability that certain pathways exist in this subject is then found by counting the number of successful trackings, divided by the total number of datasets. As a result, the final display for probabilistic tractography provides the users with only a quantitative guidance, for which the users have to determine by themselves how trustworthy the results are. More on this issue will be mentioned in a later section.
Bootstrap Probabilistic Tractography
The need for multiple DTI datasets to derive the fiber tract passing probability as stated in the previous section is certainly accompanied by an inevitable lengthening of the image-acquisition time. It is, therefore, not an attractive approach in the busy clinical environment.30 Alternatively, if multiple signal-intensity averages are used in routine image acquisition but are stored separately in the memory space, a permutation of the dataset can be performed to equivalently form multiple sets of single-average data (the so-called bootstrap approach).31 Moreover, when >6 directions are used for the diffusion-sensitizing gradients with a single signal-intensity average, it is possible to estimate the diffusion tensor fitting uncertainties, from which the probability of fiber tracts passing through certain regions can also be computed.32,33 In any case, the computations needed for probabilistic tractography are generally much more demanding than deterministic tractography algorithms.
Fiber Tracking Based on HARDI
In the presence of intravoxel fiber crossing, the inadequacy of the diffusion tensor model to fully describe multiple fiber bundles has led to the development of data-acquisition approaches called HARDI.34,35 With sufficient data sampled at various diffusion-encoding directions uniformly covering the so-called q-space, the directionality of multiple fiber tracts can be depicted by using a complex 3D shape called the orientation distribution function (Fig 5A). Just as in the case of a diffusion tensor ellipsoid, for neuroradiologists it suffices to remember that the orientation distribution function simply reflects the tendency of diffusion along various directions. This information can again be obtained from the original diffusion-weighted images on a voxel-by-voxel basis, albeit necessitating a much larger amount (sometimes >100 diffusion-encoding directions) of image data than DTI.
A, Fiber crossing within 1 image voxel can be resolved by using HARDI, where the directions of the multiple fiber bundles are depicted by orientation distribution functions magnified in the lower right corner. In the example acquired by using 1 HARDI acquisition method called Q-ball imaging as shown here, the shape of the colored orientation distribution functions shows 2 intersecting elongated objects, representing the presence of 2 major fiber tracts in the anterior body of the corpus callosum and the cingulum. B, With the orientation distribution functions obtained for all image voxels from high angular resolution diffusion imaging, fiber tracts can be connected with many possibilities. Schematic tract examples (colored thick line segments) representing 2 fiber bundles in the anterior body of the corpus callosum and the cingulum as shown in A cross each other at an oblique angle.
The directions showing the locally fastest diffusion in an orientation distribution function correspond to directions lacking substantial diffusion barriers (myelin sheath, cell membranes, organelles, proteins, etc) for the water molecules. Hence, as in the case of the longest axis of a diffusion tensor ellipsoid, directions of these local maxima are likely consistent with those of the underlying fiber bundles. In the case shown in Fig 5A, 2 major white matter tracts seem to cross each other in the voxels of interest. With all possible fiber directions included, tractography by using algorithms mentioned in the previous sections can be performed in a more comprehensive manner. Briefly speaking, all connection possibilities are considered together, and the highest directional consistency with neighboring voxels is selected to form the most probable connection (Fig 5B). The major difference from DTI tractography lies in the fact that multiple choices rather than a single choice are now available. Because each single voxel now contains multiple fiber bundles with different directions, angular consistency is relatively easier to achieve, rendering generally more fibers to be tracked.
With the presence of many fiber tracts in a large portion of the image voxels, it is not difficult to imagine that the increase in tract connection flexibility has resulted in a massive number of various tractography algorithms.36–41 Almost all algorithms demonstrate advantages over other methods in certain, albeit perhaps limited, aspects. Even though the HARDI acquisition is rather lengthy and may not be suitable for clinical practice, trying to shorten the total scanning time without much compromise in data quality has been attempted with satisfactory experience reported.42,43
Bayesian Probabilistic HARDI Tractography
Sophisticated algorithms dedicated to a combination of essentially all the major advantages in algorithms mentioned above also exist. One example is the Bayesian approach for probabilistic HARDI tractography, which uses a partial volume model containing single or multiple fiber bundles with isotropic diffusion from free water for every single voxel.44,45 With more than 50 diffusion directions encoded in a 20-minute acquisition, derivation of the uncertainty in the principal fiber direction can be done without the need for multiple acquisitions or bootstrapping.26 Following voxelwise modeling, one calculates the global connection of the tracts on a whole-brain basis under the condition of voxel-by-voxel principal fiber direction and uncertainty obtained from the diffusion imaging data (hence the name Bayesian to stand for conditional probability).45 Better accuracy is obtained with more diffusion encoding directions acquired. The result is displayed as a function of connection probability with the seed points estimated by the algorithm (Fig 6). As stated before, for probabilistic tractography in general, the users have to determine by themselves the probabilistic threshold at which the final graphic results should be shown, such that a good balance can be reached between increased false-positives versus lost tracts.
Tractography results obtained from a healthy subject, showing tracts possibly associated with the seed point placed at the lower level of the internal capsule with connection probability >0.1 (A) and 0.01 (B), respectively. Note that it is generally believed that the corticospinal tracts do not cross to the contralateral hemisphere through the corpus callosum, indicating that users have to be cautious when selecting the probability threshold to display the results. In this case, therefore, a probability threshold of 0.1 seems to be a preferred choice to 0.01. The example shown here was processed with Bayesian probabilistic tractography with 30 diffusion-encoding directions acquired.
One important characteristic of the Bayesian algorithm is that the construction of the tracts is based on an integration of the fiber direction uncertainty into the global optimization approach. When only a single fiber is modeled in a voxel, the presence of intravoxel fiber crossing would simply lead to a relatively high uncertainty for the principal fiber direction, rather than an erroneous orientation.44 Tract reconstruction based on conditional probability thus still allows a connection if the nearby neighboring voxels exhibit largely consistent fiber directions. As a consequence, stop criteria using a turning angle or FA value play a relatively minor role, with the accompanying advantages of avoiding some of the tract loss from the settings of stop criteria.36
The advancement of HARDI tractography is currently an active field of research and development, as can be evidenced by the increasing number of investigations in the literature. No matter whether one prefers the use of model-free algorithms34,35,38 or the model-based approaches,26,44,45 it is anticipated that the improved differentiation of crossing fibers in HARDI tractography will find unique neuroradiology applications that are hardly achievable by simple diffusion tensor tractography. The potentially critical role of HARDI tractography can, therefore, be expected for clinical routines in the near future, particularly if the heavy computational loads could be solved with continuous improvements in computer technology.
Limitations of Diffusion Tractography
In the authors' strictly personal opinions, the most important limitations of tractography in clinical neuroradiology lie in the nature of computational estimation based almost entirely on fiber directions in the presence of noise. There are actually 3 key words embedded in the previous statement: computational estimation, fiber direction, and noise, whose effects are obviously mutually integrated. Although each single factor could theoretically be dealt with by using proper methods specifically designed to reduce tracking errors, it is difficult to isolate the combinational influences independently. Therefore, even if the number of tractography algorithms stated above is overwhelmingly large, the fact that they are all using directional consistency as the major constraint should give one an idea of their ultimate limitations.
First, all computations are simply estimations. For any estimation method, the false-positive and false-negative rates determine the extent to which the method can be clinically useful. Unfortunately, diffusion tractography for the human brain is a case in which the results are hard to validate in vivo; hence, false-positive and false-negative rates are both unavailable. Under such circumstances, comparison with physical or mathematic phantoms27 and verification by using animal experiments,46 both having limitations to be addressed below, are probably the best one can do to test effectiveness. For the human brain in vivo, investigations on the sensitivity of the tracking results to the choice of freely adjustable parameters alone has shown that many tractography algorithms are not simple robust turn-key systems. For instance, a recent study examining the corticospinal tract concluded that the placement of seed points significantly influences the tracking results.47 This outcome is not surprising considering what we have illustrated in Fig 3B. Other reports found that the correlation of tractography results with pathologic status depends strongly on the threshold setting of FA.48,49 Moreover, in our own experience, running the same tractography algorithm with all identical parameter settings on 3 different personal computers could result in quite distinct outputs (ie, computer-dependent!), not to mention the expected confusing discrepancies in tracking results when using different software algorithms.50
The example of seed point dependency illustrated above exhibits direct difficulty in the reproducibility of tractography results. Prescribing section orientation at a slightly oblique angle may lead to rather different placement of the seed points in terms of the anatomic location, even if the region of interest is chosen carefully, particularly because the seed points are likely placed in a subvoxel coordinate. As a consequence, even if only 1 tractography algorithm is used with fixed settings of all variables, the tracking results are not free from computational imprecision. Visualization of the specific white matter pathway does not give accurate information in terms of the size or volume of the fiber bundle, either.51
Second, fiber tracking based totally on directional consistency computation risks bias in favor of connecting fibers with similar orientations. As mentioned before, highly curved fibers such as the Meyer loop may be easily missed. In addition, incidental consistency in fiber directions leads to identification of spurious tracts with no actual anatomic existence.52 This issue is further complicated by the presence of noise because of the different levels of influence on direction derivation. Estimation of the principal direction of a diffusion tensor exhibits high accuracy when the diffusion tensors exhibit high FA values and are less prone to noise influences. In contrast, for diffusion tensors whose first eigenvalue is not much larger than the second eigenvalue, the angular deviation from the true value could become rather large. Therefore, fiber tracking reproducibility was reported to be somewhat worse in the optic radiation and pyramidal tracts than in the callosal fibers.53 Likewise, the existence of a large number of studies demonstrating successful tracking of major fiber bundles such as the corpus callosum should not be regarded as the validity of tracking results of the increasingly challenging tracts such as the language pathways.54,55
Investigators having noticed the importance of estimation errors in fiber direction have proposed means to graphically depict the directional uncertainty on a voxel-by-voxel basis (Fig 7).45,56 Models applicable to identifying the limitations of tractography algorithms are also available.57 While likely useful in checking the reliability of fiber tracking results, the complexity of these tools is perhaps no less than the tractography algorithms themselves, hence substantially lowering the intention of usage for the general clinicians. It would be ideal if the uncertainty assessment could be integrated with the usage of fiber tracking algorithms as part of the training program, translating imperfect tractography into a useful clinical tool,58 which certainly entails much effort.
A, Errors in the estimation of fiber orientation can be graphically depicted on a voxel-by-voxel basis by using, for example, the double-cone diagram, in which the principal eigenvector of a diffusion tensor ellipsoid in an image voxel (upper right) is now replaced by a double cone (lower left), whose angular width represents the data-fitting uncertainty. B, An example of the double-cone map showing the fiber direction and its uncertainty simultaneously, overlaid on the original gray-scale transaxial FA map. From the magnified region (covering part of the corpus callosum and the corona radiata), it is seen that voxels showing high FA values such as the callosal fibers tend to exhibit a narrow cone width, consistent with the expectation of high angular accuracy. In contrast, uncertainty levels around 20° ∼ 30° are not uncommon even in the corona radiata for this particular image. The directional uncertainty is a strong function of the signal–to-noise ratio.
The third factor imposing limitation to the tractography algorithms reported to date lies in the difficulty of obtaining systematic verification for the human brain in vivo. Validation studies, though large in number, are mostly focused on limited aspects only and hence should not be extrapolated to the general situations unless careful attention is paid. For instance, documents demonstrating successful tractography results compared with manganese-enhanced T1-weighted imaging on animals actually validated the identification of only 1 or some specific tracts,46 instead of multiple white matter pathways as would be done with general computational estimations. The information obtained from diffusion MR tractography and manganese-enhanced T1-weighted imaging should, therefore, be treated as complementary.59 In addition, because clinical diffusion imaging with limited scanning time hardly reaches the data quality achievable in animal scans, the same algorithm applied to the human brain is anticipated to show somewhat inferior performance. As another example, most reports on HARDI tractography mainly focused on the capability of resolving fiber bundles with crossing configurations (ie, the unique capability of HARDI compared with DTI).38–40 Few have taken into account the white matter tracts exhibiting high curvature.44,60 Naturally, validation studies showing the resolving of fiber crossing on phantoms only demonstrate the limitation of the diffusion tensor model,61 rather than the real success of HARDI tractography.
All issues mentioned above indicate a clear rule in the clinical use of diffusion MR tractography: Caution is always highly warranted when performing sophisticated algorithms. Unfortunately, articles showing positive results tend to outnumber those with negative ones in the literature, giving the general audience the strong (and actually correct at least partially) impression that the noninvasive diffusion MR tractography is clinically effective and useful. Letters about negative results,55 being extremely limited in journal publication space, often do not detail possible sources of errors in a clearly reproducible manner. As a consequence, even if many pilot studies demonstrated encouraging hope,62,63 one really needs to go through the tedious procedure of a reproducibility study for an assessment of the precision. Not only are comprehensive investigations examining the clinical effectiveness of diffusion MR tractography in surgery strongly encouraged64–66 but comparative studies of the postmortem human brain specimens using a large cohort are also highly desirable.67
With continuous technical advancement in computational algorithms or via careful use of the current tools by experienced operators, precision and accuracy of diffusion MR tractography are expected to increase to a clinically satisfactory level. In such an ideal situation, one should still keep in mind that other ultimate limitations exist. Differentiation of afferent and efferent fibers is currently impossible with diffusion MR tractography.1,2,5 Ambiguity between fiber kissing and fiber crossing is another example that is hard to solve even with the HARDI data acquisition.10 Misregistrations arising from geometric distortions of the original images acquired by using the echo-planar technique have to be corrected. Furthermore, directional consistency between neighboring voxels (and hence showing successful tract connection) does not guarantee fiber integrity, even if directional consistency and fiber integrity are generally believed to be positively correlated. Nonetheless, we would like to provide some basic guidelines in the following section, to help the clinical neuroradiologists reach the level of these ultimate limitations.
Guidelines toward Applications in Clinical Neuroradiology
Always Control the Original Image Quality
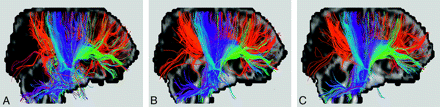
Obtaining the original images with sufficient quality is always the first step in convincing people of the validity of any processing or analysis subsequently applied to the imaging data. In diffusion MR tractography, it should be clear that the accuracy of the fiber directions is positively associated with the signal–to-noise ratio (Fig 8), which can be increased usually at the expense of scanning time or spatial resolution. Optimization of the routine protocol in a busy clinical environment is, therefore, an important prerequisite toward successful tractography.2,3 In addition, for certain tractography algorithms, there may be special requirements for the format of the original images, such as separated storage of individual acquisitions before performing signal-intensity averages68 or the preference for multiple diffusion-encoding directions over multiple signal-intensity averages.2 Before pooling the large amount of original data for the generation of beautiful 3D tractograms, it is better to make sure that the image-acquisition protocol has been adjusted in accordance with the requirements posed by the tractography algorithm.
A somewhat exaggerated comparison of the signal–to-noise ratio effects on diffusion MR tractography. Fiber bundles connected to the corpus callosum (mostly red) and the corona radiata (mostly blue) are shown in the colored tracts superimposed on a sagittal image (with the subject facing the right-handed side). The figures in A, B, and C are processed on original MR images acquired with 2, 8, and 20 signal averages, respectively. Note in A, compared with B and C, the accumulated errors in fiber orientation particularly in the inferior part of the descending tracts and the frontal portion of the callosal tracts, as well as the presence of spurious tracts.
Choose the Algorithm You Understand
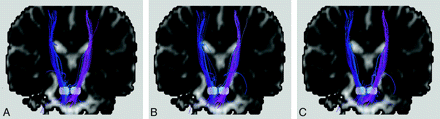
Sophisticated computational tractography algorithms, though supposedly performing better in some aspects, are not necessarily suitable for the needs in routine practice. Differences among various algorithms can be subtle (Fig 9). For example, one method may be superior to another only when the signal–to-noise ratio is above a certain level, and hence the comparison results obtained at 3T might show a reversed trend at 1.5T. Because the characteristics of algorithms vary to a great extent, choosing the one with details understood by the operators is likely the dominant factor in successful usage. When a comparison of different tractography algorithms must be made, it would be ultimately important to compare the visualization of fiber bundles with the same tracking criteria and seed points and on a standard subject or a group of subjects who are well coregistered in the same coordinate system. Under these circumstances, the performance of tractography algorithms would be minimally data- and region-of-interest-dependent, and any newly proposed algorithms could thus be tested and compared objectively.
A−C, Bilateral corticospinal tracts identified by using 3 tractography algorithms different from those used in Fig 4 but on the same healthy subject, showing subtle discrepancies among the 4 results. As in Fig 4, the 2 white regions on the coronal views are regions of interest selected to place the seed points. To avoid favoring any single algorithm, we hid the names of the algorithms. The original diffusion images were acquired with 20 signal averages to increase the signal–to-noise ratio. Tract editing was not used here to see the worst-case scenario of algorithm-dependent results.
When the choice of tractography algorithm must be based on other criteria (eg, immediate software availability, data processing time, and so forth), an equivalent alternative is to understand the algorithm one chooses. In addition to spending time studying the computational principles and playing around with freely adjustable parameter settings, trying an uncertainty test on major and minor tracts is strongly encouraged56,57,69 to have an idea of the algorithm's limitations so that one knows where to start or stop the interpretations of results. Another rough way to evaluate uncertainty is to check the left-right symmetry of reconstructed tracts when selecting a central region of interest (eg, the splenium of the corpus callosum) on images obtained from healthy subjects,70 provided that the fiber bundles do not play functional roles with known lateralization (Fig 10).71
Examination of left-right symmetry on a transaxial tractogram from a healthy subject should allow one to obtain a rough idea of the uncertainty of the tractography algorithm. In the example of callosal fibers shown here, the seed points are placed in the midsagittal plane to include the entire corpus callosum. The asymmetry here likely reflects computational errors rather than microstructural lateralization.
Perform Reproducibility Tests on Healthy Subjects
A reproducibility study using highly cooperative healthy subjects is a very important step for tractography quality control. Performing it is relatively straightforward, though admittedly rather tedious. Healthy subjects are recommended because any missing of well-known white matter tracts is unlikely to be attributed to anatomic anomaly, hence likely reflecting algorithm limitations. Through an examination of the tractography results from repeated processing, one should then identify possible sources of error and attempt a remedy. For instance, an objective algorithm with parameters all set to fixed values should allow interoperator consistency as high as intraoperator consistency, leaving the choice of regions of interest the only degree of freedom. Comparison of within-scan versus between-scan agreements is also essential in figuring out the direction of improvement in terms of either the computations or the acquisitions. In rare instances but having been encountered in the authors' experience, the same algorithm analyzing the same dataset on different computers may create discrepant results. Hence computer dependence is also among the many factors that may need to be repeatedly tested by using healthy subjects.
Bear Limitations in Mind When Applied to Clinical Practice
With all the effort spent in the preparatory work as suggested above, clinical applications on a routine basis should be largely ready. Even so, one should have a conservative attitude, keeping in mind all the technical and ultimate limitations of diffusion MR tractography. A change of the operator responsible for processing tractography or an alteration of image quality due to upgrading of the MR imaging scanner could both affect the results in a time-varying manner. Another subtle but very practical and somewhat paradoxic challenge to the neuroradiology administrator is the ever-advancing computer technology: As the calculation becomes faster and the memory space keeps increasing, the desire to obtain tractograms at a higher spatial resolution in less time will, for sure, pose strong demands on the change of the predetermined protocol. Maintaining the pre-established quality while being continuously updated in the technical progress, at the same time noticing the current (but time-varying) limitations, is certainly no less complicated than the difficult and time-consuming radiologic diagnosis of rare diseases.
Conclusions
The clinical implications and potentials of diffusion tensor MR tractography in the noninvasive investigation of white matter tracts will continue to show promise. From a visualization of pathologic alterations of the fiber bundles to presurgical evaluation of brain masses, tractography provides neuroradiologists with perhaps an elusive hope that white matter morphology can now be depicted with details exceeding the original acquisition resolution. Nevertheless, after reviewing this article, readers should agree that putting diffusion MR tractography in clinical practice with sufficient accuracy is no easy task either. With the discovery of new tracts, reasonable speculation on the technical inadequacy of the tractography algorithms remains the first healthy attitude.10 Even when obtaining fancy tractography results highly consistent with expectations, careful conservative attention in the interpretation is mandatory.
Footnotes
-
H.-W.C. and C.-Y.C. received support in part from the National Science Council under grants NSC 98-2221-E-002-095-MY3 and NSC 97-2314-B-016-028-MY3. M.-C.C. was supported by the human resource grant sponsored by the Ministry of Education to National Sun Yat-Sen University.
Indicates open access to non-subscribers at www.ajnr.org
References
- 1.↵
- 2.↵
- 3.↵
- 4.↵
- 5.↵
- 6.↵
- 7.↵
- 8.↵
- 9.↵
- 10.↵
- 11.↵
- 12.↵
- 13.↵
- 14.↵
- 15.↵
- 16.↵
- 17.↵
- 18.↵
- 19.↵
- 20.↵
- 21.↵
- 22.↵
- 23.↵
- 24.↵
- 25.↵
- 26.↵
- 27.↵
- 28.↵
- 29.↵
- 30.↵
- 31.↵
- 32.↵
- 33.↵
- 34.↵
- 35.↵
- 36.↵
- 37.↵
- 38.↵
- 39.↵
- 40.↵
- 41.↵
- 42.↵
- 43.↵
- 44.↵
- 45.↵
- 46.↵
- 47.↵
- 48.↵
- 49.↵
- 50.↵
- 51.↵
- 52.↵
- 53.↵
- 54.↵
- 55.↵
- 56.↵
- 57.↵
- 58.↵
- 59.↵
- 60.↵
- 61.↵
- 62.↵
- 63.↵
- 64.↵
- 65.↵
- 66.↵
- 67.↵
- 68.↵
- 69.↵
- 70.↵
- 71.↵
- Copyright © American Society of Neuroradiology