Abstract
BACKGROUND AND PURPOSE: Bone MR imaging techniques enable visualization of cortical bone without the need for ionizing radiation. Automated conversion of bone MR imaging to synthetic CT is highly desirable for downstream image processing and eventual clinical adoption. Given the complex anatomy and pathology of the head and neck, deep learning models are ideally suited for learning such mapping.
MATERIALS AND METHODS: This was a retrospective study of 39 pediatric and adult patients with bone MR imaging and CT examinations of the head and neck. For each patient, MR imaging and CT data sets were spatially coregistered using multiple-point affine transformation. Paired MR imaging and CT slices were generated for model training, using 4-fold cross-validation. We trained 3 different encoder-decoder models: Light_U-Net (2 million parameters) and VGG-16 U-Net (29 million parameters) without and with transfer learning. Loss functions included mean absolute error, mean squared error, and a weighted average. Performance metrics included Pearson R, mean absolute error, mean squared error, bone precision, and bone recall. We investigated model generalizability by training and validating across different conditions.
RESULTS: The Light_U-Net architecture quantitatively outperformed VGG-16 models. Mean absolute error loss resulted in higher bone precision, while mean squared error yielded higher bone recall. Performance metrics decreased when using training data captured only in a different environment but increased when local training data were augmented with those from different hospitals, vendors, or MR imaging techniques.
CONCLUSIONS: We have optimized a robust deep learning model for conversion of bone MR imaging to synthetic CT, which shows good performance and generalizability when trained on different hospitals, vendors, and MR imaging techniques. This approach shows promise for facilitating downstream image processing and adoption into clinical practice.
ABBREVIATIONS:
- DL
- deep learning
- GRE
- gradient recalled-echo
- MAE
- mean absolute error
- MSE
- mean squared error
- TE
- echo time
MR imaging is the workhorse of clinical neuroradiology, providing high tissue contrast for the evaluation of CNS structures. However, CT remains the first-line technique for rapid neurologic screening and cortical bone assessment. A novel class of MR imaging techniques uses very short TE to capture weak and short-lived proton signals from dry tissues such as cortical bone. As MR imaging hardware and software have advanced, “black-bone” MR imaging techniques have progressively improved from gradient recalled-echo (GRE) to ultrashort-TE and zero-TE approaches.1⇓-3 TE values are on the order of 1–2 ms for GRE, 50–200 ms for ultrashort-TE, and 0–25 ms for zero-TE (Online Supplemental Data). With shorter TEs, the detectable signal from cortical bone increases, scan times become faster, acoustic noise from gradient switching decreases, and resistance to motion and susceptibility artifacts increases.4-5
Bone MR imaging offers the potential for both rapid initial screening and comprehensive “one-stop-shop” imaging, without the need for ionizing radiation exposure. Thus, bone MR imaging is a promising alternative to CT for bone imaging. However, current barriers to implementation involve direct comparison of bone MR imaging and CT with regard to multiple factors, including accessibility, cost, convenience, patient awareness, clinician understanding, diagnostic accuracy, and interventional utility.6-8 A key step in facilitating clinical understanding and adoption is the automated conversion of bone MR imaging to synthetic CT-like contrast, which is highly desirable for image interpretation, 3D printing, and surgical planning applications. Conventional image-processing approaches, such as intensity thresholding, logarithmic inversion, histogram subtraction, atlas- and voxel-based techniques, have all been investigated.9⇓⇓-12 In clinical practice, these point operation–based techniques are hampered by false-negatives in the setting of undermineralization (young children or osteopenia), very thin bone (pathologic remodeling), and multiple bone-air interfaces (facial bones, skull base), as well as false-positives including other short-T2 tissues (fascia, dura, ligaments, cartilage, hardware, hemorrhage, mucoid secretions, air) and complex tissue interfaces (tumor, trauma, inflammation).
Deep learning (DL) offers a promising approach for synthetic CT generation, being already routinely used for tissue classification and image-mapping purposes. DL algorithms use multiple layers of neighborhood-based operations to derive complex information from diverse input data sets, including MR imaging signal properties, normal anatomic structures, and pathologic changes. In neuroradiology, DL for synthetic CT has been explored in adult volunteers and a few patient case series, the most frequent application being radiation therapy planning or PET attenuation correction. Despite these early studies suggesting feasibility, synthetic CT approaches have only been successful when applied to anatomically simpler regions such as the torso and extremities or normal adult skull anatomy at low spatial resolution.13-17 For most cases of head and neck clinical applications, existing synthetic CT algorithms fail due to the wide variety of normal anatomic variants and pathologic conditions. Without sufficient clinical training data and human supervision, DL-powered bone MR imaging conversion approaches show limitations similar to those of conventional processing, yielding a variety of false-negatives and false-positives.
Robust synthetic CT algorithms still have not been developed for head and neck applications, are not routinely used in clinical decision-making, and do not carry added value over source MR images interpreted by experienced radiologists. Therefore, at this time, bone MR imaging is a useful alternative to CT for diagnostic imaging but requires a radiologist’s understanding of imaging physics, head and neck anatomy, and pathologic disease processes to optimally analyze the source images. Improvement of automated synthetic CT algorithms could help address existing barriers to technology implementation by providing greater understanding for untrained radiologists and clinicians as well as facilitating downstream processing such as 3D printing and surgical navigation. Therefore, the objective of our study was to optimize a convolutional neural network algorithm for bone MR imaging conversion to synthetic CT based on our diverse data set of patients using different institutions, platforms, and bone MR imaging techniques. In particular, we sought to develop a robust DL model that would show good performance and generalizability, thus facilitating downstream image processing and adoption into clinical practice.
MATERIALS AND METHODS
Data Acquisition
This was an institutional review board–approved retrospective study with de-identified data sequentially collected from 2 institutions. The patient flowchart for study selection is described in the Online Supplemental Data. Originally, 53 patients were included with bone MR imaging and CT of the head and neck performed within a 6-month time period for bone evaluation. Following image review by a neuroradiologist with expertise in bone MR imaging, 14 patients were excluded on the basis of nondiagnostic image quality (MR imaging and/or CT) due to motion, hardware, or other artifacts. This exclusion resulted in a final data set of 39 patients: 16 patients from institution 1 and 23 patients from institution 2. Subjects spanned a broad age range (neonate to 35 years; median age, 4.5 years) with 23 (59%) male and 16 (41%) female patients. Clinical indications for imaging were suspected craniosynostosis (n = 10), genetic syndrome (n = 5), tumor (n = 4), trauma (n = 4), preoperative planning (n = 10), and postoperative follow-up (n = 6). Anatomic imaging coverage included the head, face, neck, and/or cervical spine, based on the indication. For bone MR imaging, an additional bone sequence was added to the examination on the basis of a clinical request and/or the indication for bone imaging. A variety of platforms, techniques, and field strengths were used, depending on the institution and scanner availability.
For MR, thirteen patients were scanned on Siemens Healthineers (Erlangen, Germany) platforms (3 Tesla: Magnetom Prisma, 1.5 Tesla: Magnetom Aera), and 26, on GE Healthcare (Chicago, IL) platforms (3 Tesla: Discovery MR750, MR750w). Bone MR imaging sequences were adapted from commercially available options and included 3D zero-TE, ultrashort-TE, and GRE sequences with a 20- to 30-cm FOV and 0.7- to 1-mm isotropic resolution. Sample parameters are provided in the Online Supplemental Data. Most scans were performed at 3T, with 2 scans performed at 1.5T field strength due to device-compatibility considerations. For CT, 23 examinations were performed on Siemens Healthineers platforms (Somatom Definition Flash, Somatom Definition Edge, Somatom Definition AS, Somatom Force, Somatom Sensation 64); 9, on GE Healthcare platforms (Discovery CT750 HD, Optima CT660, LightSpeed VCT); and 7 on Canon Medical Systems (Tustin, California) platforms (Aquilion ONE) using a standard multidetector technique (age-adjusted radiation dose, 0.5- to 1-mm section thickness, bone reconstruction kernel).
Image Coregistration and Preprocessing
The goal of the image-processing pipeline (Online Supplemental Data) was to generate a diverse set of spatially aligned bone MR imaging and CT pairs for neural network training. A neuroradiologist with experience in bone MR imaging coregistered all MR imaging and CT images on the basis of key anatomic landmarks and inspected the final matched image sets for quality assurance. First, multiple-point affine transformation of MR imaging to CT data was performed in OsiriX MD (http://www.osirix-viewer.com) to yield coregistered 3D volumes. All remaining image-preprocessing steps were implemented in Matlab (MathWorks). The image volumes were resampled to achieve isotropic resolution in all dimensions and then were divided into paired 2D MR imaging and CT slices in axial, coronal, and sagittal planes. While synthesizing only axial CT views may be sufficient for many applications, we were interested in deriving the largest and most diverse training set possible. Each image pair was masked and cropped to disregard irrelevant background artifacts during training. Masks were created by binarizing the CT image (using Otsu’s method to find the ideal threshold18) and finding the largest convex area in the binary image. The same convex mask was also applied to the paired MR images. Images were cropped to the smallest possible square containing and centering the masked content.
Finally, each section was resized to the resolution required for neural network input. The resulting images were saved with an 8-bit gray-scale depth based on the entire dynamic range for MR imaging slices and bone window/level for CT slices. On average, this pipeline generated 550 MR imaging/CT pairs per patient (approximately 22,000 image slices total). Additionally, we artificially augmented our training data by randomly flipping (horizontally or vertically), rotating (by <10°), or cropping (by <10%) image pairs during training.
We note here that masking the MR image based on a registered CT image would not be possible in a real-world scenario (because CT would not be available). However, we found this approach to work much more robustly, which was necessary to automate the masking pipeline, given the large amount of training slices. Our goal was to have clean training data. Inference based on a nonmasked MR imaging is still possible.
Neural Network Architectures
We tested 3 encoder-decoder networks based on U-Net models.19 For the first model, we built a lightweight baseline model (Light_U-Net) based on the original U-Net architecture but decreasing the number of filters (channels) for each block, for total of ∼2 million trainable parameters. We further changed the filter size of the transposed convolutions from 2 × 2 to 3 × 3 so that the decoder path exactly mirrored the encoder path, avoiding the need to crop the filter responses in the skip connections. The output layer was reduced to a single channel with sigmoid activation function, allowing the model to produce a gray-scale image rather than a binary segmentation mask. For the second model, we used the well-established VGG-16 convolutional neural network architecture20 for the encoder path and mirrored it for the decoder path. The resulting model had a larger number of filters and filter blocks, resulting in ∼29 million total trainable parameters. This enabled us to use transfer-learning as a third model variation, VGG-16 U-Net transfer-learning, in which filter weights in the encoder path were initialized with values learned from the public ImageNet (https://image-net.org/index) data set, in which a large variety of annotated objects were classified from >14 million conventional color photographs21 (Online Supplemental Data).
Model Implementation
All DL models were implemented in Python (Python Software Foundation) using the TensorFlow library (www.tensorflow.org) with the Keras interface (Massachusetts Institute of Technology). All experiments were run on a high-performance computing cluster using either a NVIDIA Tesla V100 or NVIDIA Tesla P100 GPU (Nvidia, Santa Clara, California). The input to the model was a single-channel gray-scale bone MR image with a resolution of 224 × 224 pixels to match the fixed resolution of the VGG-16 architecture. Each 3 × 3 convolutional layer was followed by a batch-normalization layer and a ReLU activation layer. The VGG-16 U-Net architecture, which was originally designed for color images, required a 3-channel image input, so the gray-scale image was repeated across all 3 channels. Because the encoder path was an exact copy of the original VGG-16 architecture, its 3 × 3 convolutional layers were not followed by a batch-normalization layer but only had an ReLU activation. For the decoder path, batch normalization was still added after every 3 × 3 convolutional layer. For both networks, the synthetic CT image was produced via a 1 × 1 convolutional layer with a sigmoid activation, creating a continuous gray-scale image on the interval of 0–1 and a resolution of 224 × 224 pixels. All models were optimized with stochastic gradient descent using the Adam method22 with default parameters and a batch size of 128 images. Network weights were initialized randomly, except for the VGG-16 U-Net transfer learning variant, in which weights in the encoder path were pretrained on ImageNet. No weights were frozen during optimization.
Loss Functions
We experimented with optimizing 3 different loss functions: mean absolute error (MAE, also called L1 loss), mean squared error (MSE, also called L2 loss), and a weighted sum of both:

where N is the total number of image pairs in the training set and α is a coefficient that was selected empirically as 4.4, resulting in approximately equal contribution of L1 and L2 to the total loss.
Model Training and Validation
Because intrapatient image slices are visibly correlated with each other compared with interpatient slices, we trained and evaluated our models on data from separate patients. For every experiment, we performed a patient-level 4-fold cross-validation, with each model trained on three-quarters of the patients and then tested on the remaining quarter. Reported results were aggregated across all 4 models.
Because neural network optimization is stochastic in nature (random initialization and random batching), training on the same data set multiple times may result in a different model convergence. We, therefore, repeated each 4-fold cross-validation experiment 10 times and reported average performance and 95% confidence intervals across the 10 independent runs.
Neural network models additionally require an internal validation set to prevent overfitting. For this purpose, a random 15% of slices from the training data were held out during training. After each training epoch, we computed the internal validation loss and stopped training the model once that validation loss had not decreased for at least 5 epochs. We selected the model weights with the smallest internal validation loss up to that point.
Performance Metrics
Global performance metrics were calculated pixel-wise across the image data sets and included MAE, MSE, and the Pearson correlation coefficient R. To express MAE and MSE in terms of Hounsfield units, we rescaled the neural network output on the basis of a window width of 2000 HU. In addition, we quantified the degree of bone overlap between ground truth CT and synthetic CT by thresholding both into binary bone maps. Given a threshold t, we defined bone precision, bone recall (sensitivity), and bone Dice score as

Thresholding was done on a grid of thresholds ranging from 40% gray level to 70% gray level (Online Supplemental Data). We report the average precision, recall (sensitivity), and Dice score across all thresholds.
Model Generalizability
Because our full data set contained images acquired at different hospitals, as well as using difference imaging vendors and bone MR imaging techniques, we conducted a series of experiments to evaluate how well model performance generalizes across all these different dimensions. All models were based on Light_U-Net with MAE loss. For each test set, baseline model performance was computed using patient-based 4-fold cross-validation with a training set from the same data subset (vendor, hospital, or MR imaging technique). These baseline results were compared with a model trained only on data from a separate subset, as well as a model trained on augmented data including both the current and separate subsets (again with 4-fold cross-validation).
RESULTS
Model Architectures and Loss Functions
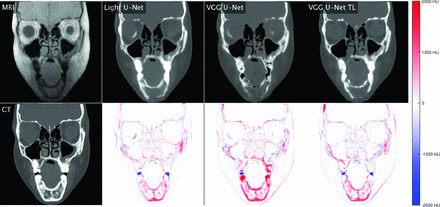
Performance comparison of the various neural network models and 3 loss functions is summarized in the Table, with visual comparison of model results in Fig 1 and loss functions in Fig 2. Results are based on 10 repetitions of a patient-based 4-fold cross-validation among the 16 patients from institution 1, which contained the best quality and most curated data. Among all model architectures, Light_U-Net achieved the lowest test MAE and MSE when trained with MAE and MSE loss, respectively. Light_U-Net models also achieved the highest correlation coefficients across the board. When trained on the mixture loss, Light_U-Net also achieved a lower test MAE and MSE than both VGG U-Net and VGG U-Net transfer learning. Adding transfer learning to VGG U-Net tended to increase the test performance, though differences between VGG U-Net and VGG U-Net transfer learning were not always significant.
Four-fold cross-validation results for different model and loss combinationsa
Comparison of different encoder-decoder models. The first column shows real MR imaging and real CT. Subsequent columns show synthetic CTs generated by Light_U-Net, VGG U-Net, and VGG U-Net transfer learning, as well as pixel-wise difference maps between synthetic CT and real CT. Red indicates that synthetic CT is darker than real CT; blue, synthetic CT is brighter than real CT (Refer online version for colors).
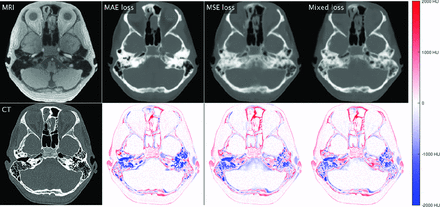
Comparison of different loss functions using a Light_U-Net model. The first column shows real MR imaging and real CT. Subsequent columns show synthetic CTs generated when using a loss function based on MAE, MSE, and a mixed combination, as well as pixel-wise difference maps between synthetic CT and real CT. Red indicates that synthetic CT is darker than real CT; blue, synthetic CT is brighter than real CT (Refer online version for colors).
When we compared loss functions, models trained on MAE loss naturally achieved a lower validation MAE than those trained on MSE loss and vice versa, with the mixture loss falling in-between. MAE loss achieved a significantly higher mean bone precision across all network architectures. Visually, the synthetic CT images showed sharper edge contrast with crisper bone detail. In addition, relatively fewer pixels were assigned bone density (white signal) on CT, indicating higher specificity, a lower false-positive rate, and higher false-negative rate for bone. MSE tended to achieve a higher mean bone recall (sensitivity) with various network architectures, though the differences were not statistically significant. Visually, the synthetic CT images showed margins that were more blurry and more homogenized bone detail. In addition, relatively more pixels were assigned bone density (white signal) on CT, indicating a higher sensitivity, higher false-positive rate, and lower false-negative rate for bone. In general, MAE loss tended to undercall bone, and MSE loss tended to overcall bone, with the mixture loss producing intermediate image effects.
Overall, the Light_U-Net architecture models outperform or tie other models in all metrics, with difference loss functions allowing adjustment among higher bone precision, recall, or overlap (Dice score). Additional examples of synthetic CT images in axial, coronal, and sagittal views are provided in the Online Supplemental Data. When reviewed by expert neuroradiologists, the computationally optimized model (Light_U-Net, MAE) yielded visibly superior results compared with previously reported synthetic CT algorithms (eg, conventional logarithmic inversion and vendor-provided processing tools). For example, our algorithm enabled delineation of bone microstructure in typically false-negative areas of thin bone (facial bones, skull base, remodeled bone). In addition, our algorithm better excluded false-positive areas such as the fascia and mucoid secretions. Finally, the synthetic CT images showed distinction of nonbone tissues, including soft tissue, fat, and air, that was comparable with the true CT.
Model Generalizability
Results of generalizability experiments across different hospitals, vendors, and bone MR imaging techniques are summarized in the Online Supplemental Data. Training on additional data from a different hospital, vendor, or technique significantly improves performance in terms of MAE, MSE, and the correlation coefficient for all conditions, even when the added patients are few in number compared with the reference data set. Conversely, when one trains models on purely separate hospitals, vendors, or MR imaging techniques, performance significantly decreases across the board.
DISCUSSION
Model Architectures and Loss Functions
With regard to DL architecture, there are 2 classes of models that are suitable for image-to-image translation: encoder-decoder networks and conditional generative adversarial networks. Generative adversarial networks have the distinct advantage of learning to synthesize realistic-looking images when paired images from the source and target domain (eg, coregistered MR imaging and CT slices) are unavailable during training.23-26 In the presence of paired CT/MR imaging training data, recent experiments suggest that encoder-decoder networks tend to outperform generative adversarial networks in the CT/MR imaging domain in terms of MAE, MSE, and other metrics.27 We selected the U-Net architecture in particular because its skip connections between each encoder and decoder layer allow precise spatial information from the MR imaging to be propagated to the synthetic CT.
While transfer learning has been traditionally considered helpful when training large models for tasks with relatively small data sets (as is often the case for medical imaging), our study suggests that for MR imaging-to-CT image synthesis, smaller models with fewer training parameters may be more suitable. This result is supported by recent systematic studies that found that the transfer accuracy (specifically with models pretrained on ImageNet) is very sensitive to how exactly the pretraining was done.28-30 For example, many common forms of regularization may increase ImageNet accuracy but are less suited for transfer learning. An alternative transfer learning approach for future experiments could include finding a related image-translation task for which paired training data are available on a large scale. In general, if more training data are available, larger models may still be able to perform better for this task.
In terms of error minimization, low loss based on pixel-level statistics does not ensure a visually convincing and spatially accurate image rendering. We attempted to numerically quantify synthetic CT image quality by measuring bone precision, recall, and Dice scores on the basis of multiple gray-level thresholds. In addition, clinical assessment of synthetic CT images was performed by a neuroradiologist with expertise in bone MR imaging. Both numerically and visually, there were competing trade-offs in MAE-versus-MSE loss, and these trends persisted across all network architectures. This persistence can be because MAE error is computationally more tolerant of abrupt intensity changes between neighboring pixels, allowing small local errors and less bulk density assignment of bone. Therefore, MAE loss achieves higher precision, higher specificity, a lower false-positive rate, and higher a false-negative rate for bone. Visually, this results in a high-contrast image with sharply defined edges and a tendency to undercall bone. On the other hand, MSE loss penalizes individual outliers more heavily and so enforces a more universally balanced error. Therefore, MSE loss achieves higher recall (sensitivity), a higher false-positive rate, and a lower false-negative rate for bone. Visually, these findings result in a smoother and more regularized image with bulk density assignment to larger areas and a tendency to overcall bone. Using a mixture loss allows a balance among these competing factors, suggesting that the weighting coefficient α could be titrated depending on the clinical use case.
As previously mentioned, prior synthetic CT articles have used conventional or DL-based approaches in anatomically simpler regions, including the normal adult head, torso, and extremities, for nondiagnostic applications, including radiation therapy planning and PET attenuation and correction.13-17 More recent work has also described conventional or DL-powered approaches to synthetic CT using other MR imaging sequences such as GRE, T1, and T2.31,32 The physics of these sequences is inherently less sensitive to cortical bone so that postprocessing approaches are destined to be less accurate. Indeed, the sample synthetic CT output from these articles is low-resolution and insufficient for diagnostic radiology use.
Review of the computationally optimized model (Light_U-Net, MAE) by expert neuroradiologists showed clear potential clinical value over existing conventional and DL algorithms. Our synthetic CT algorithm visibly recaptured bone microstructure in areas pushing the limits of the MR imaging technique, generated fewer false-negative and false-positive bone assignments, and enabled distinction among nonbone tissues. Given that the network architectures and loss functions we used are similar to those described in prior DL studies, our improved results are best attributed to the use of real-world clinical data.
Advancement of clinical implementation will need to include large-scale systematic human reviews of DL algorithms to quantify the usefulness for diagnostic evaluation and interventional planning. At our institution, we are conducting a noninferiority trial of bone MR imaging versus CT, with CT representing the criterion standard technique or ground truth. Expert radiologists are independently evaluating CT, bone MR imaging, and synthetic CT images (Light_U-Net, MAE) to provide numeric scores (0–10) for visibility of key anatomic landmarks (calvaria, sutures, fontanelles, orbits, nose, jaw, teeth, paranasal sinuses, skull base, temporal bone, and cervical spine). For the patients analyzed in this study, CT landmark mean ratings ranged from 9.4 (SD , 0.52) for the calvaria to 9.1 (SD , 0.91) for temporal bone. For MR imaging, the highest rated landmark was also the calvaria (mean, 9.0 [SD , 0.86]) and the lowest was the temporal bone (mean , 7.2 [SD 1.39]). For synthetic CT, the highest rated landmark was the calvaria (mean , 8.1 [SD , 0.92]) and the lowest was the paranasal sinus (mean, 6.8 [SD , 2.31]). These preliminary data suggest that landmark visibility on bone MR imaging and synthetic CT are slightly less than on real CT but sufficient to make most clinical diagnoses.
Furthermore, we are comparing the suitability of CT, bone MR imaging, and synthetic CT data sets for 3D anatomic modeling and virtual surgical planning. Biomedical engineers are processing imaging volumes via bone segmentation, mesh triangulation, and surface generation. Conventional anatomic modeling pipelines use CT with density thresholding to identify bone. Therefore, source bone MR imaging with multiple dark structures is difficult and time-consuming to manually segment. In our experience, synthetic CT algorithms greatly facilitate 3D processing workflow, though as noted by radiologists, anatomic accuracy is less in challenging areas such as facial bones and skull base. For each patient, we coregister image data to calculate a matrix of the reference CT surface and spatial deviation Δ of the nearest point on the test surface (synthetic CT), displayed as a color heat map. We can then calculate statistical metrics over the entire point cloud (mean, range, SD, interquartile range). Based on the surgical accuracy criteria, we can also compute the percentage of data falling within the clinically acceptable tolerance interval Δ = (–2 mm, +2 mm). For the patients analyzed in this study, 86% (SD, 0.18) of all MR imaging surface data falls within ±2 mm of coregistered CT surface data. The largest areas of deviation are attributed to missing MR imaging data around regions of hardware and difficult-to-segment anatomic areas, which will guide further investigation.33-36
Future comparative effectiveness studies will need to account for the relative risks and benefits of clinical workflow, ionizing radiation exposure, examination duration, anesthesia requirements, diagnostic quality, and treatment outcomes. For example, bone MR imaging represents a key alternative for at-risk patients in whom radiation exposure must be minimized or eliminated, ie, children, pregnant women, and patients with cancer-predisposition syndromes. In such patients, CT dose reduction can yield poor image quality below a certain dose threshold. Therefore, ultra-low-dose CT versus no-dose bone MR imaging may yield a more realistic and equitable image comparison.7,33-36
Model Generalizability
In general, DL approaches benefit from larger and more broadly representative training data. This study is limited by the relatively small sample size of 39 patients, which, nevertheless, represents the largest documented database of paired bone MR imaging and CT examinations in clinical patients. Because referring patterns can vary across clinicians and institutions, we chose to include all available head and neck imaging cases to maximize the volume and diversity of the data set. Our study cohort includes varied patient ages, backgrounds, and disease processes with generalizable real-world imaging data, including motion and artifacts. We standardized the preprocessing and conversion of these volumetric data sets into a unique image repository of approximately 22,000 2D paired MR imaging and CT image slices. It would be advisable for multiple institutions interested in bone MR imaging and CT to create a multicenter consortium that can establish best practices with regard to clinical referrals, bone MR imaging techniques, image preprocessing, data sharing, and model development to further increase the available volume and scope of training data. As enrollment numbers increase, it may be possible to develop algorithms tailored to specific clinical indications. This collaborative effort will help elevate collaborations and democratize access among radiologists, clinicians, and patients worldwide.
Our cross-validation experiments evaluated the impact of different hospitals, vendors, and bone MR imaging techniques on model generalizability. These generalizability experiments showed that training on an augmented data set that includes a different hospital, vendor, or technique significantly improves model performance. Conversely, when one trains models only on disparate data sets, performance significantly decreases across the board. Taken together, these results suggest that blindly applying a model trained only on an outside data set can be dangerous due to inherent data variations, but augmenting a local model with additional data sets can boost overall performance. These are key considerations for any institution looking to practically implement bone MR imaging and synthetic CT. Future computational work will involve further model optimization and customization of problem-specific loss functions. We are also considering processing input data in patches, which would permit assembly of higher-resolution output images than our current model.37-41
Having established a robust DL pipeline with good performance and generalizability, we hope to facilitate adoption into clinical practice. At our institution, we are already seeing early promise for diagnostic and interventional applications. With larger clinical training sets, continued enhancement of synthetic CT algorithms will improve understanding among untrained radiologists and clinicians and streamline downstream processing for 3D printing and surgical navigation. Further technical advancements could even augment diagnostic value over source MR images, as suggested by the ability to reconstruct bone microstructure approaching MR imaging super-resolution. As synthetic CT algorithms become more robust and accessible, they may be increasingly accepted for clinical decision-making in head and neck imaging. True clinical validation will require comparative effectiveness research across different clinical use cases and multiple iterations of human expert input to guide selection and implementation of optimal algorithms.
CONCLUSIONS
We have optimized a DL model for conversion of bone MR imaging to synthetic CT in the head and neck on the basis of a patient data set inclusive of diverse demographics and clinical use cases. Our unique database consists of 39 paired bone MR imaging and CT examinations, scanned at 2 different institutions with varying MR imaging vendors and techniques. The Light_U-Net model outperformed more complex VGG U-Net models, even after the use of transfer learning. Selection of loss function on the basis of MAE resulted in better bone precision, while MSE tended to provide better bone recall. Performance metrics for a given model decreased when using training data captured only in a different environment and increased when local training data were augmented with those from different hospitals, vendors, and techniques. By establishing a robust DL-powered synthetic CT algorithm with good performance and generalizability, we hope to elevate the applicability of bone MR imaging with downstream image-processing and adoption into clinical practice.
Acknowledgments
We would like to thank Houchun Harry Hu, PhD, Mark Smith, MS, Aiming Lu, PhD, and Bhavani Selvaraj, MS, for their scientific expertise and collaboration. We would also like to thank Lisa Martin, MD, Diana Rodriguez, MD, Jeremy Jones, MD, Charles Elmaraghy, MD, Eric Sribnick, MD, Ibrahim Khansa, MD, and Gregory Pearson, MD, for their clinical expertise.
Footnotes
This work was supported by the American Society of Head & Neck Radiology Core Curriculum Fund William N. Hanafee Research Grant, Siemens/Radiological Society of North America Research Scholar Grant, RSCH1804, and the Society for Pediatric Radiology Pilot Award.
Disclosures: Mai-Lan Ho—RELATED: Grant: RSNA, SPR, ASHNR Comments: RSNA Research Scholar Grant, SPR Pilot Award, ASHNR William N. Hanafee Grant.* Support for Travel to Meetings for the Study or Other Purposes: RSNA, SPR, ASHNR Comments: RSNA Research Scholar Grant, SPR Pilot Award, ASHNR William N. Hanafee Grant.* UNRELATED—Royalties: McGraw-Hill Comments: Author, Neuroradiology Signs. *Money paid to the institution.
References
- Received March 30, 2021.
- Accepted after revision June 13, 2022.
- © 2022 by American Journal of Neuroradiology