Abstract
BACKGROUND AND PURPOSE: The complex MR imaging appearance of glioblastoma is a function of underlying histopathologic heterogeneity. A better understanding of these correlations, particularly the influence of infiltrating glioma cells and vasogenic edema on T2 and diffusivity signal in nonenhancing areas, has important implications in the management of these patients. With localized biopsies, the objective of this study was to generate a model capable of predicting cellularity at each voxel within an entire tumor volume as a function of signal intensity, thus providing a means of quantifying tumor infiltration into surrounding brain tissue.
MATERIALS AND METHODS: Ninety-one localized biopsies were obtained from 36 patients with glioblastoma. Signal intensities corresponding to these samples were derived from T1-postcontrast subtraction, T2-FLAIR, and ADC sequences by using an automated coregistration algorithm. Cell density was calculated for each specimen by using an automated cell-counting algorithm. Signal intensity was plotted against cell density for each MR image.
RESULTS: T2-FLAIR (r = −0.61) and ADC (r = −0.63) sequences were inversely correlated with cell density. T1-postcontrast (r = 0.69) subtraction was directly correlated with cell density. Combining these relationships yielded a multiparametric model with improved correlation (r = 0.74), suggesting that each sequence offers different and complementary information.
CONCLUSIONS: Using localized biopsies, we have generated a model that illustrates a quantitative and significant relationship between MR signal and cell density. Projecting this relationship over the entire tumor volume allows mapping of the intratumoral heterogeneity in both the contrast-enhancing tumor core and nonenhancing margins of glioblastoma and may be used to guide extended surgical resection, localized biopsies, and radiation field mapping.
ABBREVIATIONS:
- CE
- contrast-enhancing
- HPF
- high-power field
- NE
- nonenhancing
Glioblastoma is a complex malignancy characterized by heterogeneous radiographic and histopathologic features. This intratumoral heterogeneity, combined with the diffuse infiltration of glioblastoma, renders correlations between imaging and underlying tissue critical for better understanding of tumor behavior.1 Stereotactically localized biopsies allow this comparison of local radiologic characteristics across multiple sequences with histopathologic or molecular analysis and provide a means for understanding underlying tissue characteristics that lead to macroscopic appearances on MR imaging.1⇓⇓–4 However to date, histopathologic-radiographic correlations have been limited by the following: 1) the surgical challenge of obtaining stereotactically localized biopsies with reliable accuracy, 2) the maintenance of spatial fidelity across multiple MR images, and 3) the application of quantitative histopathologic analysis.
Radiographically, glioblastoma typically appears as a contrast-enhancing (CE) mass with surrounding nonenhancing (NE) tissue marked by abnormal T2-FLAIR and DWI-derived ADC signals.5⇓–7 These heterogeneous MR signal characteristics in the NE area are known to represent a combination of vasogenic edema and infiltrating glioma cells8⇓–10; however, correlating these MR imaging abnormalities with underlying histopathology remains challenging. Nonetheless, there has been much effort with a variety of MR images to draw such radiologic-histologic correlations to better understand the tumor environment,1⇓⇓–4 which is critical for refining radiation treatment fields, assessing response to therapy, and evaluating progression.
Early studies have suggested that tumor cellularity as a surrogate for infiltration correlates with decreased apparent diffusivity,3,10⇓⇓–13 reduced T2-FLAIR hyperintensity,14,15 and increased gadolinium-related T1 signal.16,17 More recent effort has combined these changes on multiple MR images to help distinguish glioma cell infiltration and edema. Akbari et al14 used a machine-learning algorithm to identify several multiparametric features that spatially colocalized with areas of future radiographic recurrence, with the implication that these regions likely harbor underlying infiltrative tumor. However, this study did not confirm the proposed radiographic features with histologic data. Hu et al18 incorporated multiple MR imaging textural features with histologic data to identify areas of high tumor content for targeting diagnostic biopsies. The resulting model was able to sort tissue into 2 categories of high and low tumor content. Together, these initial studies suggested that important imaging patterns exist amid the complex signal changes within the peritumoral region and that careful analysis may yield correlates to tumor infiltration.
We hypothesized that quantitative measures of tissue cellularity from localized biopsies will correlate with voxel-level signal on multiple MR images and will provide more informative maps of tumor infiltration at the margins of glioblastomas. To this end, a fully automated cell counting and image coregistration pipeline was developed to eliminate subjective bias and improve reliability. With this approach, the objective of this study was to generate a model capable of predicting cellularity at each voxel within an entire tumor volume as a function of signal intensity, thus providing a means of quantifying tumor infiltration into surrounding brain tissue.
Materials and Methods
Patient Selection
A retrospective review was performed on our data base of adult patients with MR imaging–localized biopsies obtained during open surgical resection for glioma at Columbia University Medical Center between January 2012 and January 2015. Patient selection was limited to those with newly diagnosed glioblastoma (World Health Organization grade IV) resected by 1 of 2 attending neurosurgeons (M.B.S., J.N.B.), who had complete preoperative MR imaging available for analysis (ADC, T2-FLAIR, and T1-weighted precontrast and postcontrast sequences). Patients were excluded if their localized samples were determined to be inadequate for any for any of the following reasons: poor neuronavigational registration, excessive MR imaging motion artifacts, errors in the automated screen capture, or problems with histologic processing pipeline.
Image Acquisition
All imaging was performed on a 3T MR imaging system (Signa; GE Healthcare, Milwaukee, Wisconsin) by using an 8-channel head-array coil (Signa HDxt; GE Healthcare). The following sequences were assessed for each patient: pre- and postcontrast T1-weighted, T2-FLAIR, and DWI. Pre- and postcontrast volumetric acquisition was performed with a T1-weighted 3D inversion recovery fast-spoiled gradient-recalled sequence with the following parameters: TI = 450 ms, TR = 10.2 ms, TE = 4.2 ms, flip angle α = 13°, FOV = 250 mm; matrix = 256 × 256, section thickness = 1.2 mm. The total scan time was approximately 4 minutes 15 seconds. Postcontrast images were acquired by using intravenous gadobenate dimeglumine (MultiHance; Bracco Diagnostics, Princeton, New Jersey) at a dose of 0.2 mL/kg. The time between injection and postcontrast imaging was approximately 5 minutes. Axial T2-FLAIR was performed with the following parameters: TR/TE = 9500/127 ms, TI = 2250 ms, section thickness = 5 mm with no gap, FOV = 225 mm. DWI was performed with a single-shot, spin-echo, echo-planar sequence with the following parameters: TR/TE =7000/73 ms, FOV = 220 mm, matrix = 128 × 192, section thickness = 5 mm with no gap, bandwidth = 1953 Hz/pixel with b-values of 0 and 1000 s/mm2. ADC maps were generated in FuncTool software (GE Healthcare) for the quantitative determination of mean diffusivity measurements.
Image Preprocessing
Raw image data from T1-precontrast, T2-FLAIR, and ADC maps were aligned to the volumetric T1-postcontrast sequence by using the FMRIB Linear Image Registration Tool (FLIRT; http://www.fmrib.ox.ac.uk/).19,20 The linear affine transformation algorithm was implemented by using 12 df, trilinear interpolation, and a mutual information cost function. Each sequence was further processed with a histogram normalization algorithm to standardize intensity values among patients. The algorithm is based on a method previously described that maximizes the cross-correlation between cumulative histogram distribution functions of each input volume with a reference standard.21 A mean reference template was created for each sequence by pooling data from the study subjects, each set to a mean intensity of 0 and an SD of 1. T1-postcontrast subtraction images were obtained by subtracting coregistration-normalized T1-precontrast volumes from T1-postcontrast volumes.
Biopsy Acquisition
All tissue sampling was performed within the normal surgical plan when it posed no additional risk to the patient. Samples were taken from the CE tumor core in all cases and the NE region around the tumor in some cases. Sampling of the NE, T2-FLAIR hyperintense region was permissible in 2 scenarios: 1) when the surgical trajectory required dissection through brain parenchyma en route to the tumor, and 2) when resection of tissue lateral or superficial to the CE region was part of the surgical goal. Biopsies were obtained before tumor debulking to maximize the spatial fidelity of localization. Stereotactic guidance was provided by a volumetric T1-weighted postcontrast sequence on a neuronavigation system (Brainlab Curve; Brainlab, Feldkirchen, Germany). The location of the biopsy was recorded by screen capture of the surgical navigation system at the time of tissue sampling.
Biopsy Coregistration
A custom fully automated 2D-to-3D coregistration algorithm was developed to convert the crosshairs from the neuronavigation system screenshot image into an MR imaging coordinate (On-line Fig 1). The algorithm is implemented in the following steps: First, a cropped subimage of the biopsy crosshair in the axial orientation is obtained. Subsequently, a 2D convolution is applied to a single axial section from the original MR imaging by using the cropped axial subimage as a convolution kernel. The process is iterated over a number of subimage scales (30%–200% zoom) and MR imaging sections by using a grid search technique. The maximum from each resulting cross-correlation response matrix is recorded, and the parameters are updated until convergence is achieved. A final 2D convolution is applied to the image by using a kernel reflecting a mask of the expected crosshair, thus localizing the point of interest. With this coordinate as the center point, all voxels within a spheric ROI radius of 1 mm are recorded for each coregistered sequence. The final intensity value used for analysis from each biopsy point is a Gaussian-weighted mean of the voxels within the spheric ROI.
Histopathologic Processing and Analysis
The samples were divided into 2 pieces in the operating suite. One piece was immediately flash frozen in liquid nitrogen for future genetic studies. The second piece was fixed in 10% (volume/volume) formalin and infiltrated with low-temperature paraffin for histologic analysis. Five-micrometer sections were stained with hematoxylin-eosin. The slides were then scanned and digitalized to BigTIFF files (http://bigtiff.org/) at ×40 magnification by using a Leica SCN400 system (Leica Biosystems, Wetzlar, Germany). The high-resolution image was acquired at 40,000 pixels per centimeter (2.5 × 10−5 cm per pixel).
Whole-Slide Cell Counting
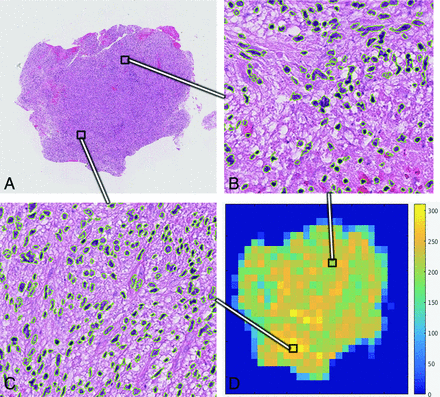
A fully automated cell-counting algorithm was developed by using a convolutional neural network (Fig 1 and On-line Fig 2). The algorithm was trained to count the number of nuclei present in a single high-power field (HPF), defined by a 2.25 × 2.25 mm square (900 × 900 pixels). Subsequently, the neural network was iteratively processed through tiles of HPFs until the entire H&E slide was counted (Fig 1B–D). HPFs at the border of the tissue specimen (eg, only a fraction of the field contained cells) were excluded from analysis.
Whole-cell counting. A, Digitized, low-power magnification view of a single H&E-stained slide. Two representative 400× fields from this single tissue specimen of relatively lower (B) and higher (C) cell density illustrate the tissue heterogeneity present at a microscopic level. Stained cellular nuclei identified by the automated counting algorithm are outlined in green. D, The “heat map” demonstrates distribution of cell density at the level of a HPF throughout the tissue sample.
The neural network was trained to classify the center pixel of a 21 × 21 × 3 RGB color input into 1 of 3 categories: cellular nuclei, densely staining artifacts mimicking cellular nuclei, and background staining. After a hyperparameter grid search, the final neural network architecture consisted of 5 convolutional layers of 5 × 5 filters (with increasing channel filter depths of 20, 40, 60, 80, and 120) followed by a final fully connected layer. Rectified linear activation functions were used after each convolutional layer, and a final softmax loss function was used to classify each input image. The network was trained on 5000 annotated examples of each class and validated on separate 1000 ground truth examples. A final validation classification error of 3.8% was achieved after training for 100 epochs.
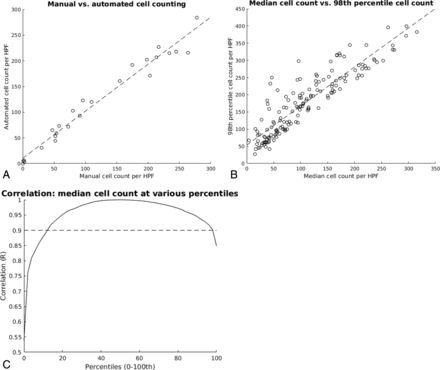
As a final test set validation, 25 high-power fields were chosen at random. Each field was manually inspected to determine the number of nuclei present. The same field was then evaluated by the automated cell-counting algorithm. The human reviewer was blinded to the final total cell count obtained by the automated method. A high correlation was observed between the automated algorithm and the manual cell counts as determined by a Pearson coefficient (r = 0.984, Fig 2A).
Cell-counting statistics. A, Comparison between manual and automated cell counts for 25 high-power fields of various cellular densities. Correlation is high (r = 0.984), suggesting that the automated algorithm accurately reflects manual counts. B, For each biopsy sample, the median cell density of all HPFs is compared with that of the 98th percentile. A relatively strong linear correlation is preserved (r = 0.901), suggesting that the 98th percentile cell density simply represents a linear translation of the median cell density. C, Correlation analysis is repeated for all percentiles (0–100). With the exception of extreme values, most percentiles retain a strong linear correlation (r > 90%) with the median cell density.
Statistical Analysis
For each biopsy, MR signal intensity (ADC, T2-FLAIR, and T1-weighted postcontrast) was correlated against cell density per HPF. Single variable linear regression analysis and Pearson correlation coefficients were determined. In addition, a multiple linear regression model was created as defined by:
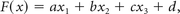 where the predicted cellularity, F(x), was a linear function of signal intensity on ADC, T2-FLAIR, and T1-weighted postcontrast sequences (x1, x2, and x3, respectively).
where the predicted cellularity, F(x), was a linear function of signal intensity on ADC, T2-FLAIR, and T1-weighted postcontrast sequences (x1, x2, and x3, respectively).
To quantify the effects of potential biopsy misregistration, we repeated the above linear correlations iteratively by replacing the original spheric ROI with concentric shells of voxels at an increasing distance from the biopsy center. The resulting changes to the correlation of the model with increasing distance were plotted and fit with a sigmoidal regression curve. All statistical and image analyses were performed by using Matlab 2015b (MathWorks, Natick, Massachusetts).
Results
Patient Population
Thirty-six patients were included in this study, of whom 15 were male and 21 were female. The mean patient age at the operation was 65.2 years (range, 24–88 years). Eight patients were excluded due to poor neuronavigational registration, excessive motion artifacts, or errors in radiographic or tissue processing. A total of 91 localized biopsies were analyzed (median, 2 samples per patient; range, 1–6 samples). Each biopsy point was coregistered individually on ADC, T2-FLAIR, and T1-weighted postcontrast subtraction sequences, yielding 273 sets of data points for analysis.
Cell Density
Ninety-one H&E slides, 30,578 HPFs (mean, 336 HPFs per sample), were analyzed.
The median cell density was 98 nuclei per HPF (range, 4–296 nuclei) among all biopsies. The median was used as a representative measure for cell density in an H&E slide, given the presence of both outlying high- and low-density HPFs in almost all tissue samples (Fig 1B, -C). Other percentile cell densities were also considered, but it was shown that these represented an approximately linear translation of the median within any given H&E slide (Fig 2B, -C). For example, the 98th percentile cell density of any given H&E slide was directly linearly correlated to the median cell count (r = 0.901), displaced by approximately 50 additional nuclei/HPFs. Thus, although the following analysis was evaluated with the median cell density, the linear nature of the relationship between signal intensity and cellularity would remain valid across most cell density percentiles.
Correlation between Signal Intensity and Cell Density
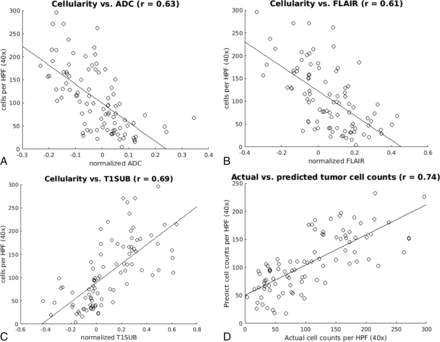
Scatterplots were used to assess the relationship between MR signal intensity and cellularity on multiple sequences (Fig 3). The correlation between ADC and cellularity was r = 0.63; the correlation between T2-FLAIR and cellularity was r = 0.61; and the correlation between T1-weighted postcontrast subtraction and cellularity was r = 0.69. A multivariable linear regression was performed to assess the predictive power of the combined MR images. The Table shows the results of this analysis. The combined model correlation improves to r = 0.74 with a model variance of R2 = 0.55.
Cell count versus MR signal intensity. Scatterplots demonstrate median cell density as a function of signal intensity on ADC (A), T2-FLAIR (B), and T1-postcontrast subtraction sequences (C) correlated by using single-variable regression analysis. The linear regression and Pearson correlation (r) were significant (P < .05) for all 3 sequences. D, The scatterplot shows the actual and predicted cell counts as estimated by combining all 3 imaging modalities in a multiple-variable regression model.
Multivariate linear regression model coefficients
Effects of Possible Misregistration
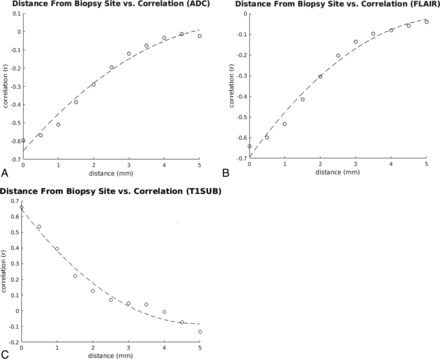
The ability to correlate voxel-level signal intensities and histopathology is contingent on accurate biopsy colocalization. To estimate the detrimental effect of possible misregistrations by our automated coregistration algorithm, we compared the relationship between MR signal intensity at varying degrees of registration error. The spheric ROI used for the model correlation was systematically replaced with voxels displaced at 0.5-mm increments from the center up to 5 mm of displacement. For example, the first ROI included all voxels located 0.5 mm from the center, the second ROI included all voxels 1.0 mm from the center, and so on, resulting in a series of concentric spheric shells. The correlations between signal intensity and these projected biopsy locations are shown in Fig 4. As expected, the correlation decreased as a function of distance from the original biopsy site. After 5 mm of displacement, near-zero correlation was observed for ADC and T2-FLAIR sequences, suggesting that our registration algorithm was accurate to >5-mm spatial resolution. Of note, increasing sampling distance beyond this threshold for T1-weighted postcontrast subtraction sequences inverts the correlation from positive to negative. This outcome is likely because beyond a certain distance, voxels originally within the CE tumor are now predominantly in the NE region, while voxels within the NE portion of the tumor likely begin to incorporate portions of CE tumor, thus inverting the expected relationship between enhancement and cellularity.
Correlation versus distance from the biopsy. Scatterplots demonstrate the correlation between cell density and signal intensity for each MR image (ADC, T2-FLAIR, T1-postcontrast subtraction) obtained by taking the mean of concentric spheric shells of voxels at an increasing distance from the original biopsy point. Notably, the correlations drop to 0 at a radius of approximately 5 mm (∼10 voxels), providing an estimate of the spatial accuracy of the biopsy location. T1SUB indicates T1-subtraction.
Cell Density Map
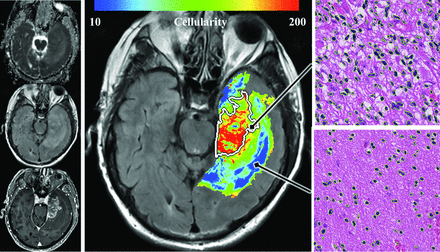
Estimates for cellularity were projected at each voxel within the tumor volume by using the multivariable linear regression model and coregistered MR imaging sequences (Fig 5). Given that all biopsies were obtained within the boundaries of T2-FLAIR signal abnormality, the projected model estimates are limited to these regions.
Whole-tumor model overlay. Estimated cellularity by applying the multiple regression model on a voxelwise basis across the tumor. The model is derived from linear regression by using ADC, T2-FLAIR, and T1-postcontrast sequences shown in the inset on the left. In the right panels, corresponding biopsy specimens (400× magnification, H&E stained sections) are shown from 2 regions obtained on the same section, highlighting the considerable variation in cellularity in and around the region of contrast enhancement (demarcated by a white outline).
Discussion
In this study, we describe a voxel-level multiparametric MR imaging model of glioblastoma cellularity derived from the radiologic and histologic features of radiographically localized biopsies. We found that tumor cellularity is inversely correlated with ADC and T2-FLAIR signal and directly correlated with T1-weighted postcontrast signal. A multiparametric linear regression model based on these correlations captured approximately 54% of the variance in cellularity within the tumor. Projection of these estimates throughout the tumor volume provided a noninvasive map of glioblastoma cellularity, suggesting a means of visualizing infiltrative margins that may be derived from routine standard imaging. Clinically, this model may be used to guide extended surgical resection, localized biopsies, and/or radiation field mapping. Furthermore, it can be used as an outcome measure for tracking progression and response to treatment.
Cellularity was inversely correlated to ADC signal; this correlation is consistent with the notion that water diffusion is restricted in hypercellular neoplastic environments.11,12,22⇓–24 This concept would suggest reduced ADC signal from frank tumor tissue in the area of CE region, but also from infiltrating tumor in the surrounding NE area. Our results are concordant with prior studies using localized biopsies in the CE area, which found an inverse relationship between ADC and cellularity in the CE core.3,25 However, studies that used this technique in the peritumoral region yielded contradictory results. Sadeghi et al3 failed to identify any significant relationship of ADC and cellularity in the peritumoral region; however, this study consisted largely of lower grade gliomas and the peritumoral region was defined histologically rather than radiologically. Conversely, the postmortem analysis of ex vivo tissue samples of LaViolette et al10 found a significant inverse relationship between ADC and cellularity in areas of tissue coregistered to peritumoral T2-FLAIR hyperintensity. In summary, the mixed results and differences in techniques yielded no strong conclusions but, together with our results, suggested that ADC offers some information in characterizing the peritumoral region. However, additional study is necessary to understand precisely how tumor histology is represented by ADC.
Cellularity was also inversely correlated with T2-FLAIR signal intensity. This correlation confirms earlier hypotheses of variations in T2-FLAIR hyperintensity. Specifically, areas of high cell density result in an intermediate intensity signal, while tissue with greater proportions of vasogenic edema is more uniformly hyperintense.15 This distinction was previously suggested by Akbari et al,14 in which a mild loss in T2-FLAIR signal was found to be a significant predictor of cellular infiltration, leading to recurrent glioblastoma. Our results are the first to confirm this observation with histologic sampling, to our knowledge.
Conversely, cellularity was directly correlated with T1-shortening on gadolinium-enhanced sequences. This finding is in line with evidence of the perivascular migration of malignant cells, disruption of endothelial tight junctions, and subsequent leakage of gadolinium.16,17,26,27 Notably, instead of a bimodal distribution of T1 signal intensity corresponding to CE-versus-NE tumor, a broad range of MR imaging intensity values was observed. These observations corresponded to a similarly continuous broad distribution of cellularity. These findings suggest that even within apparently similar tumor compartments, marked histologic heterogeneity is present.
To account for the possible effects of intrinsic T1 shortening as may be seen with intratumoral blood products or dystrophic mineralization, we performed an analysis by using images obtained by subtracting T1-weighted precontrast and postcontrast data. Compared with results with native T1-weighted postcontrast images, the overall correlation in both single and multivariable models is slightly superior (On-line Fig 3). One potential explanation is the reduced confounding influence of intrinsic T1 shortening when using subtraction images, though no significant hemorrhage or dystrophic mineralization was identified on histopathologic evaluation of localized biopsy samples. A second possible source of improved correlation is that subtraction images facilitate an internal reference for intensity normalization so that local magnetic field inhomogeneities or other artifacts present on both pre- and postcontrast imaging would be reduced.
A multiple linear regression model combining these individual relationships improved overall correlation from 61%–69% to 74%. This improvement confirms that these MR images contribute different and complementary information about the magnetic properties of the tumor environment. Specifically, our model is informed by tumor effects on Brownian motion of water, tissue heterogeneity in vasogenic edema, and the degree of tumor-induced blood-brain barrier breakdown.
Multiparametric modeling has been previously leveraged to investigate the imaging surrogates of peritumoral infiltration. Akbari et al14 used imaging evidence of tumor recurrence in lieu of histologic markers for cellularity to identify MR imaging features suggestive of glioblastoma infiltration, yielding a predictive model for recurrence. Hu et al18 expanded on this technique with histologic correlation, by using textural features extracted from multiple MR images to identify regions of tumor-rich biopsy targets (defined as ≥80% tumor content) in both the CE and NE regions. Our investigation builds on the described previous work for multiparametric analysis by implementing quantitative analysis of cellularity in the localized biopsies from both CE and NE regions. The use of this continuous variable provides a predictive model of underlying tissue heterogeneity.
Furthermore, this study relates radiologic and histologic characteristics at a more granular level, estimating cellularity on a voxel-by-voxel basis. This feature necessitates accurate and precise biopsy acquisition, coregistration of multiple imaging sequences, and quantitative histologic analysis of tissue samples. This effort remains nontrivial, with several potential sources of error, including the following: neuronavigational registration to the patient's cranial features, intraoperative brain shift, initial conversion of the biopsy image capture to 3D MR imaging space, differences in acquisition matrices and patient motion, and analysis of whole-tissue specimens for full representation of their heterogeneity.2,28
Measures were taken intraoperatively, and downstream processing was fully automated to minimize these potential sources of errors. By using changes in the model correlation as a reference for deleterious effects secondary to possible misregistration, we demonstrate that gradually displacing our original biopsy coordinate by >0.5 mm causes an overall loss of signal correlation with tumor cellularity. This outcome suggests that the coordinates derived from the neuronavigation system screen capture correspond well to the true location of tissue sampling. Furthermore, the cell-counting algorithm provided unbiased measures of cell density over the entirety of these heterogeneous tissue samples.
This proposed multivariable model reflects simple and intuitive signal-intensity responses to varying tissue cellularity; however, ultimately only 55% of the variance in observed tumor cellularity is captured. The variance not accounted for by our model may stem from several sources: The first is localization error, which includes shifts in tissue position during an operation, errors in registration between the MR imaging and neuronavigational system, and errors in registration in each patient's multimodal MRIs. An additional source of unexplained variance comes from the MR imaging intensity of each voxel being affected by many physical and physiologic factors (eg, cell size and shape, water volume, diffusion anisotropy, and so forth). Although these factors are affected by tumor cellularity, a one-to-one relationship between cellularity and MR imaging intensity of any given sequence does not exist. While a multivariable approach, in part, reduces this phenomenon by minimizing the effects of uncorrelated noise, the proposed model is ultimately limited to linear relationships between signal intensity and cellularity, unable to account for the more spatially complex radiologic features of glioblastoma. The ability to model nonlinear behavior in signal intensity may be improved in future iterations by using machine-learning algorithms to implement both supervised and unsupervised feature detection.
While cellularity provided a simple metric for radiographic-histologic correlation, future studies would benefit from using localized biopsies to analyze additional histologic characteristics and molecular markers to capture other, more sophisticated features of biologic heterogeneity in glioblastoma. In fact, the use of machine-learning algorithms to implement both supervised and unsupervised feature detection may allow the model to account for potential complex and nonlinear radiographic-histologic relationships. The incorporation of additional physiologic imaging parameters such as dynamic susceptibility contrast perfusion, diffusion tensor imaging, and resting-state blood oxygen level–dependent imaging would likely further improve model prediction. Finally, the model may generalize to other types of gliomas (eg, low-grade gliomas); however, any model predictions beyond glioblastoma would require a validation similar to that performed in the current study.
Conclusions
This study demonstrates a significant correlation between voxel-level signal intensity and cell density in glioblastoma by single-variable regression as well as a more powerful multiparametric predictive model. Thus, with a precise and rigorous analysis pipeline, we have affirmed the feasibility of meaningful quantitative analysis at the voxel-level between signal intensity and localized biopsies. In addition, the multiparametric model proposed in this study provides a means to noninvasively map cell density at the infiltrative margins of glioblastoma. This characterization, supplemented by other histopathologic features, may be used to guide extended surgical resection, localized biopsy, and/or radiation field mapping, with significant implications for patient management.
Footnotes
Peter D. Chang and Hani R. Malone contributed equally to this work.
Disclosures: Adam M. Sonabend—UNRELATED: Employment: Columbia University; Grants/Grants Pending: National Institutes of Health, Comments: I have a DP5 National Institutes of Health grant from the Office of the Director, National Institutes of Health. Lawrence H. Schwartz—OTHER RELATIONSHIPS: consulting or advisory role company: Novartis, GlaxoSmithKline; research funding company: Eli Lilly, Astellas Pharma, Merck Sharp & Dohme, Pfizer, Boehringer Ingelheim; patent licensing for Varian Medical Systems. Peter Canoll—RELATED: Grant: James S. McDonnell Foundation, Irving Institute for Clinical and Translational Research: Pilot Research Award, Comments: These grants supported the analysis of the localized biopsies.
This work was supported by the James S. McDonnell Foundation and a Collaborative and Multidisciplinary Pilot Research Award from the Irving Institute for Clinical and Translational Research, Columbia University Medical Center.
REFERENCES
- Received June 13, 2016.
- Accepted after revision December 9, 2016.
- © 2017 by American Journal of Neuroradiology