Abstract
BACKGROUND AND PURPOSE: Perfusion MR imaging measures of relative CBV can distinguish recurrent tumor from posttreatment radiation effects in high-grade gliomas. Currently, relative CBV measurement requires normalization based on user-defined reference tissues. A recently proposed method of relative CBV standardization eliminates the need for user input. This study compares the predictive performance of relative CBV standardization against relative CBV normalization for quantifying recurrent tumor burden in high-grade gliomas relative to posttreatment radiation effects.
MATERIALS AND METHODS: We recruited 38 previously treated patients with high-grade gliomas (World Health Organization grades III or IV) undergoing surgical re-resection for new contrast-enhancing lesions concerning for recurrent tumor versus posttreatment radiation effects. We recovered 112 image-localized biopsies and quantified the percentage of histologic tumor content versus posttreatment radiation effects for each sample. We measured spatially matched normalized and standardized relative CBV metrics (mean, median) and fractional tumor burden for each biopsy. We compared relative CBV performance to predict tumor content, including the Pearson correlation (r), against histologic tumor content (0%–100%) and the receiver operating characteristic area under the curve for predicting high-versus-low tumor content using binary histologic cutoffs (≥50%; ≥80% tumor).
RESULTS: Across relative CBV metrics, fractional tumor burden showed the highest correlations with tumor content (0%–100%) for normalized (r = 0.63, P < .001) and standardized (r = 0.66, P < .001) values. With binary cutoffs (ie, ≥50%; ≥80% tumor), predictive accuracies were similar for both standardized and normalized metrics and across relative CBV metrics. Median relative CBV achieved the highest area under the curve (normalized = 0.87, standardized = 0.86) for predicting ≥50% tumor, while fractional tumor burden achieved the highest area under the curve (normalized = 0.77, standardized = 0.80) for predicting ≥80% tumor.
CONCLUSIONS: Standardization of relative CBV achieves similar performance compared with normalized relative CBV and offers an important step toward workflow optimization and consensus methodology.
ABBREVIATIONS:
- AUC
- area under the curve
- FTB
- fractional tumor burden
- GBM
- glioblastoma multiforme
- NAWM
- normal-appearing white matter
- PTRE
- posttreatment radiation effects
- rCBV
- relative CBV
- ROC
- receiver operating characteristic
Conventional contrast-enhanced MR imaging guides response assessment for essentially all patients with high-grade gliomas and clinical trials worldwide. Despite this widespread use, diagnostic challenges remain. In particular, nontumoral posttreatment radiation effects (PTRE), namely pseudoprogression and radiation necrosis, can exactly mimic tumor recurrence on contrast-enhanced MR imaging.1⇓⇓-4 While tumor recurrence signals treatment failure, PTRE represents a positive response to treatment with a good prognosis. This distinction can be further complicated by the histologic admixture between tumor and PTRE, which can impact both diagnosis and prognostication, depending on the relative histologic burden of each entity.5⇓⇓-8 In the case of surgical biopsy, regional heterogeneity and resulting sampling errors can negatively affect the diagnostic confirmation of tumor versus PTRE, as well as the adequacy of tumor content for molecular and genomic profiling.9 These issues underscore the importance of improving image-based response assessment, particularly in the context of intratumoral heterogeneity.
During the past decade, DSC-MR imaging has emerged as a clinically valuable and accessible tool to distinguish tumor recurrence from PTRE.10 A continually expanding body of literature has shown how DSC-MR imaging measures of relative cerebral blood volume (rCBV) are significantly higher in tumor compared with PTRE.1,10⇓⇓⇓-14 These studies have spurred the development of proposed guidelines for interpretation and have helped promote clinical adoption and accessibility across an increasing number of clinical practices.15 At the same time, this expansion has brought to light issues of interinstitutional variability in DSC-MR imaging methodology, which can affect how rCBV is clinically measured and interpreted.10,16,17 This variability has motivated the effort to develop consensus recommendations on DSC-MR imaging acquisition and postprocessing that can help standardize clinical practice across institutions.15
Of the various methodologic factors that can impact rCBV measurement (eg, software modeling,18,19 preload dose,16,17,20,21 pulse sequence parameters16,17), the process of rCBV normalization is arguably the most fundamental.22⇓-24 While quantitative measurements of rCBV are not possible given the poor reliability of arterial input functions on DSC-MR imaging and unknown voxelwise contrast agent T2* relaxivity, rCBV normalization provides a means of semiquantification against internal reference tissue in each patient. By convention, users define ROIs within the contralateral normal-appearing white matter (NAWM) and/or normal gray matter.13,20 These user-defined inputs are susceptible to variability (eg, size and location of ROIs, vascularity of selected reference tissues), which can lead to substantial variability in rCBV measurements.20 The technique of standardization has been proposed to eliminate the need for user-defined input and thereby reduce rCBV variabilities.22 This process transforms rCBV maps to a standardized intensity scale, without the need for reference ROIs, and has been shown to increase the consistency in rCBV measurements across time and patients.23
While standardized rCBV thresholds have also been proposed to distinguish tumor from PTRE, these have not been prospectively evaluated.11 The use of standardized rCBV is currently not widely accepted due, in part, to a need for tissue-validation studies. To address this issue, we undertook this study and hypothesized that standardized rCBV values will demonstrate similar, if not better, predictive performance in distinguishing tumor from PTRE compared with normalized rCBV. Thus, this study will compare the predictive accuracy of standardized and normalized rCBV values using a dataset of image-localized biopsies and coregistered DSC-MR imaging perfusion maps in a cohort of patients with recurrent high-grade gliomas, to distinguish and quantify the spatial heterogeneity of tumor recurrence relative to PTRE.
MATERIALS AND METHODS
Patients
We prospectively recruited previously treated (surgery, chemoradiation therapy) patients with World Health Organization grades III and IV primary high-grade gliomas, undergoing preoperative imaging for surgical re-resection of newly developed contrast-enhancing lesions identified on surveillance MR imaging. Recruitment was from 2007 to 2018. Patients were recruited from 2 institutions (Mayo Clinic and Barrow Neurological Institute). We included patients who previously underwent standard adjuvant chemoradiation therapy per the Stupp protocol.25 All our patients were diagnosed on the basis of clinical standards at the time of enrollment. We also recorded the molecular status for isocitrate dehydrogenase (IDH) 1/2 mutations and the codeletion status of 1p/19q for those patients clinically diagnosed on the basis of the revised 2016 World Health Organization classification criteria.26 The institutional review boards of each institution approved our study, and written and informed consent was obtained from each subject.
MR Imaging Protocol, Parametric Maps, and Image Coregistration
Conventional MR Imaging and Acquisition Conditions.
We acquired 3T MR imaging (Sigma HDx, GE Healthcare, Milwaukee, Wisconsin; Ingenia, Philips Healthcare, Best, Netherlands; Magnetom Skyra, Siemens, Erlangen Germany) within 1 day before stereotactic surgery. Conventional MR imaging and advanced MR imaging parameters have been detailed previously.27 Briefly, we acquired precontrast and postcontrast T1-weighted echo-spoiled gradient-echo inversion-recovery images (TI/TR/TE = 300/6.8/2.8 ms; matrix = 320 × 224; FOV = 26 cm; thickness = 2 mm). We acquired postcontrast T1-weighted images after completing DSC perfusion MR imaging following a total Gd-DTPA dosage of 0.15 mmol/kg as previously described.5,13,20,27⇓-29
Preoperative DSC-MR Imaging Protocol.
We used a 3T MR imaging system for all patients. After antecubital fossa or forearm intravenous catheter placement, we administered 0.1 mmol/kg of Gd-DTPA contrast agent (gadodiamide or gadobenate dimeglumine) preload dose 6 minutes before the DSC-MR imaging acquisition to minimize T1-weighted leakage effects.20,21,30,31 During the DSC-MR imaging acquisition, we administered a 0.05-mmol/kg bolus at 3–5 mL/s. The DSC-MR imaging sequence parameters were the following: gradient-echo EPI with TR/TE/flip angle = 1500–2000 ms/20 ms/60°; FOV = 24 × 24 cm; matrix = 128 × 128; NEX = one; 5-mm sections; no interslice gap. These parameters yielded an in-plane spatial resolution of 1.8 × 1.8 mm and voxel volumes of 16 mm3. The total Gd-DTPA dose was 0.15 mmol/kg of body weight.
rCBV and Fractional Tumor Burden Map Generation
Overview of Image Segmentation, Coregistration, and Parametric Map Generation.
For each patient, we used IB RadTech (Version 2.0.1259; Imaging Biometrics, Elm Grove, Wisconsin) to perform the following: 1) segment all T1-weighted postcontrast enhancing lesion voxels; 2) generate both normalized rCBV and standardized rCBV maps; 3) coregister all rCBV maps to respective T1-weighted postcontrast images; and 4) perform rCBV thresholding to generate separate fractional tumor burden (FTB) maps from the respective normalized and standardized rCBV maps. (Note because absolute rCBV values are not determined, the map is a relative CBV map; ie, the values are interpreted relative to other values throughout the brain in a given patient.) IB RadTech automates the previously described manual functions that were detailed for FTB map generation.5 Voxelwise rCBV was calculated on the basis of well-established methods.20,21,30 In short, after excluding the first 4 time points of each DSC-MR imaging series to reach a steady-state, we normalized the signal intensity–to-baseline and converted the dynamic series to change the relaxation rate across time [ΔR2*(t)] for the entire brain. We generated rCBV maps by integrating the area under the ΔR2*(t) curve, ending at the time point 40 seconds after the nadir signal intensity of the first-pass bolus. All rCBV values were corrected for residual T1-/T2-/T2*-weighted leakage effects using the well-established Boxerman-Schmainda-Weisskoff algorithm.20,21,30
Normalized rCBV and FTB Maps.
We generated normalized rCBV and normalized FTB maps using IB RadTech, based on a previously published workflow,5 which we also summarize in On-line Fig 1. The generation of normalized rCBV required a user to manually select 5 × 5 mm ROIs from the contralateral NAWM. To reduce the potential for user-associated variability, we developed criteria to guide the selection of uniform ROI locations within the frontal and parietal lobes, which are illustrated in On-line Fig 1.5,13,20 These ROI locations were chosen specifically to help minimize potential volume averaging from adjacent cortical/deep gray matter, adjacent ventricles, and adjacent large surface vessels along the cortex. All NAWM ROIs were selected by a single board-certified neuroradiologist (L.S.H.) with >15 years of experience. For normalized FTB, we used the normalized rCBV threshold of 1.0 (tumor > 1.0; PTRE ≤ 1.0), which has previously been established through image-localized biopsy and spatially matched histopathologic correlation.5,13,20 FTB is defined as the percentage of tumor voxels within a region with rCBV > 1.0 (On-line Fig 2). To assess potential user-dependent variability, we assigned another board-certified neuroradiologist (J.M.H.), also with >15 years of experience, to separately guide placement of NAWM ROIs, which were used to generate a separate set of normalized rCBV and normalized FTB maps. We used these data to assess interrater reliability and evaluate potential differences in predictive accuracy, which are presented in (On-line Table).
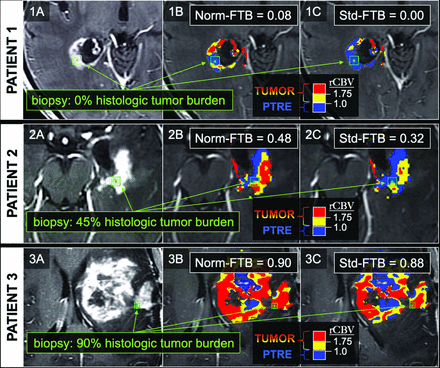
FTB maps in 3 separate biopsy cases correlating with low, medium, and high histologic tumor content. The 3 rows correspond to 3 separate patients and 3 separate biopsy locations, as shown by the green square ROIs on the anatomic postcontrast images in the far-left column (1A, 1B, 1C). The middle and far right columns show the biopsy locations (green square ROIs) in relation to the normalized (Norm) FTB (1B, 2B, 3B) and standardized (Std) FTB (1C, 2C, 3C) maps. On the FTB maps, blue corresponds to predicted PTRE regions with low rCBV ≤1.0. The yellow (1.75 ≥ rCBV >1.0) and red (rCBV >1.75) correspond to predicted tumor regions. For this study, FTB was defined as the percentage of both yellow and red voxels relative to all voxels within an ROI.
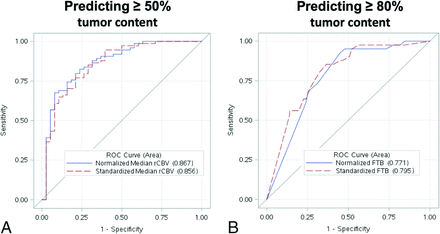
ROC analyses of the top-performing rCBV metrics for predicting ≥ 50% histologic tumor content (A) and ≥ 80% histologic tumor content (B). A, ROC curve for predicting ≥50% histologic tumor content using normalized (blue curve) and standardized (red dotted curve) median rCBV. B, ROC curve for predicting ≥80% histologic tumor content using normalized (blue curve) and standardized (red dotted curve) FTB. The standardized metrics show similar performance compared with corresponding normalized metrics.
Patient demographics and distribution of recurrent tumors by pathology and grade
Standardized rCBV and FTB Maps.
Standardized rCBV maps were generated in IB RadTech on the basis of the previously described algorithm that transforms rCBV intensities to an initial standard scale of 0–50,000.11,22,23 This transformation is achieved in 2 steps: a training step that is executed only once and a transformation step that is executed for each new input rCBV image. The training step was previously performed,22 resulting in a calibration rule (historically referred to as the “standardization step”) in which standard rCBV intensity values [μ’j(Lk)] for prechosen percentile landmarks (Lk) were determined (On-line Fig 2A). Subsequently, each new input rCBV map can be transformed to the standardized space (ie, calibrated) so that the input rCBV intensity histogram is deformed to match the standard histogram. As shown (On-line Fig 2B), this step usually results in a nonlinear intensity transformation for a given image. However, the relationship between tissue intensities is maintained, and intensity comparisons can be made using the standardized images.32 For comparison purposes (to normalized FTB), the standardized values were divided by 3575, the value shown to match an rCBV value of 1. The standardized rCBV value of 3575 was found, in a separate study, to accurately distinguish tumor from treatment effect, which was validated with spatially correlated biopsy tissue and coregistered standardized rCBV values.11 The threshold standardized rCBV of 1.0 was used (tumor > 1.0; PTRE ≤ 1.0) to define FTB as the percentage of tumor voxels within a region with rCBV > 1.0.
Acquisition of Surgical Tissue, Criteria for Histologic Diagnosis, and Estimation of Histologic Tumor Fraction
We collected multiple image-localized biopsies for each patient during surgical re-resection of MR imaging enhancing lesions for suspected recurrent disease.5,13,20,29 In short, each neurosurgeon collected an average of 2–3 biopsy specimens from spatially distinct regions within different poles of each MR imaging enhancing lesion, following the smallest possible diameter craniotomy to minimize brain shift. Biopsy target locations were based on clinical feasibility (eg, accessibility of the target site, overlying vessels, eloquent brain). The neurosurgeons recorded biopsy locations via screen capture to allow subsequent coregistration with DSC and conventional MR imaging datasets. The neurosurgeon visually validated stereotactic imaging locations with corresponding intracranial anatomic landmarks, such as vascular structures and ventricle margins, before recording specimen locations as previously described.27,28 We fixed all surgical tissue specimens in 10% formalin, embedded in paraffin, sectioned (10-μ), and hematoxylin-eosin-stained per standard diagnostic protocol at our institutions. The presence and quantity of recurrent tumor and/or PTRE elements were determined by expert neuropathology review (J.M.E.) as previously described.5,7,33 In short, for all submitted surgical tissue, we estimated the tumor burden (ie, tumor content; range, 0%–100%) relative to nonneoplastic features, on the basis of relative fractions of neoplastic features versus treatment effects, as previously described. Areas of tumor and PTRE were marked on each slide, grossly measured, microscopically re-evaluated, and used to determine the percentage of tumor/PTRE for each biopsy specimen. PTRE features included paucicellularity; scattered, rare, or no atypical cells; lack of mitotic figures except in inflammatory cells; preponderance of reactive cells including astrocytes (gemistocytes), microglia, and macrophages; and vascular hyaline fibrosis. Necrosis, often circumscribed in nonneoplastic parenchyma, if present, was considered specific for PTRE. Features of tumor recurrence included cellular sheets and/or nests of atypical cells often with mitotic figures. If necrosis was present, it was a minor component of the cellular tumor rather than parenchyma. The finding of fewer atypical cells in a linear infiltrative configuration in parenchyma without prominent reactive changes was also classified as recurrence.5,7
Extraction of Regional rCBV Metrics from Image-Localized Biopsy Locations.
All FTB and rCBV maps were exported and processed through our image-analysis pipeline, which includes coregistration of all image datasets to a standardized resolution (1.17 × 1.17 × 3 mm) as previously described.27,28 We generated 5 × 5 voxel ROIs at the stereotactic coordinates for each biopsy location for feature extraction, and the location of each ROI was verified by expert neuroradiology review (J.M.H. and L.S.H.) at the time of image analysis. We measured FTB and base statistical metrics (eg, mean, mode, SD) from each ROI for both standardized and normalized rCBV maps. This step enabled subsequent statistical analysis between spatially matched rCBV metrics and corresponding histologic quantification of tumor burden for each respective image-localized biopsy specimen.
Statistical Analysis and Predictive Accuracy.
We estimated Pearson correlation coefficients (r) between spatially matched rCBV metrics (ie, FTB, mean and median rCBV) and histologic tumor content (range, 1%–100%) from corresponding biopsies for both standardized and normalized maps. We also performed receiver operating characteristic (ROC) analysis and determined the area under the curve (AUC) for standardized and normalized rCBV metrics to predict tumor content for each sample using 2 different histologic thresholds (≥50% tumor, ≥80% tumor). We compared Pearson correlations and the AUC for standardized-versus-normalized rCBV metrics. We defined statistical significance as P < .05.
RESULTS
Subject Population and Clinical Data
We recruited a total of 38 patients with recurrent high-grade gliomas undergoing surgical re-resection and collected a total of 134 image-localized biopsy samples. We had to exclude 22 biopsy samples from subsequent image processing and correlative analysis, due to either insufficient biopsy material (n = 1), biopsy location near the skull base causing susceptibility artifacts (n = 1), biopsy location from nonenhancing parenchyma (n = 3), biopsy locations along a thin cystic wall/resection cavity that caused ROI overlap with nonparenchymal CSF-containing structures (eg, fluid-filled cyst, central necrotic core, resection cavity, n = 8), biopsy location adjacent to large vessels (n = 4), or inadequate coregistration of image datasets with biopsy location (n = 5). This exclusion allowed us to include 112 specimens from 38 patients for final analysis. Thirty-five patients were recruited from Barrow Neurological Institute and scanned on the GE MR imaging system. Two patients were recruited from Barrow Neurological Institute and scanned on the Phillips MR imaging system. One patient was recruited from Mayo Clinic and scanned on the Siemens MR imaging system. We could not identify significant differences in MR imaging image quality or rCBV measurements between scanners or institutions.
All patients had previously undergone standard adjuvant chemotherapy and radiation therapy.25 One patient was recruited twice for 2 separate operations (both for recurrent glioblastoma multiforme [GBM]) that were 8 months apart. The number of biopsies ranged from 1 to 6 per patient. Patient demographics and clinical data are summarized in Table 1. Thirty patients were recruited before 2016 and were clinically/pathologically diagnosed using the World Health Organization criteria before the 2016 recommendations. Thus, IDH and 1p/19q codeletion status were unknown in these patients. The remaining 8 patients (all with recurrent GBM) were diagnosed clinically/pathologically using the revised 2016 World Health Organization classification criteria: Five recurrent GBM tumors were IDH wild-type, and 3 recurrent GBM tumors were IDH-mutant (1p/19q intact).
Predictive Accuracies and Correlations between rCBV-Based Metrics and Histologic Tumor Content
All correlations and predictive accuracies are summarized in Table 2. Pearson correlations with histologic tumor content as a continuous variable (0%–100%) were highest with the FTB metric, showing similar coefficients for normalized FTB (r = 0.63, P < .001) and standardized FTB (r = 0.66, P < .001). Figure 1 shows 3 separate cases correlating FTB maps with spatially matched histologic tumor content from corresponding biopsies. As shown in Table 2, correlations among mean and median rCBV were slightly higher for standardized compared with normalized metrics. When we distinguished histologic tumor content based on binary cutoffs (ie, ≥50% tumor, ≥80% tumor), predictive accuracies were similar for both standardized and normalized metrics (Table 2). Median rCBV achieved the highest AUC (normalized = 0.87, standardized = 0.86) for predicting at least 50% tumor. Meanwhile, FTB achieved the highest AUC (normalized = 0.77, standardized = 0.80) for predicting at least 80% tumor (Table 2). The ROC analyses are also summarized in Fig 2. The remaining ROC analyses are shown in On-line Figs 3 and 4. We also evaluated interrater reliability by comparing normalized rCBV and normalized FTB metrics from a second user defining NAWM regions. As shown in the On-line Table, the intraclass correlation between the main user and second user–defined normalized FTB and mean and median rCBV equaled 0.89, 0.92, and 0.92, respectively, based on a 2-way model with absolute agreement. These intraclass correlations suggest strong agreement between the 2 users.
Summary of predictive performance for normalized and standardized rCBV metrics
DISCUSSION
As a primary goal of this study, we sought to determine the impact of rCBV standardization on predictive accuracy, compared with the conventional method of rCBV normalization. Our data show that standardized rCBV metrics can achieve predictive performance that is similar (if not slightly superior) to that of normalized rCBV. While standardized and normalized rCBV are both reflective of tissue vascular volume fraction, as developed, the standardization method sets values above the 99.8th percentile of each rCBV map to the maximum standardized value.22 This threshold eliminates unrealistic upper bound landmarks computed from spurious signals that might skew the histogram of the prestandardized image data. Presumably these values correspond to large vessels, either near cortical margins or within large feeding or draining vessels that course through the enhancing lesions themselves. While these vessels potentially skew normalized rCBV to overestimate tumor content,5,11 standardized rCBV would be less susceptible to these biases. Nevertheless, we found that tumor standardized rCBV values fell within the middle of the standardized range. Standardization of rCBV also eliminates user-defined input (ie, normal ROIs), shown to improve inter- and intrapatient consistency and repeatability of rCBV compared with manually defined normal ROIs.23 It is possible that these improvements in variance and reproducibility may help bolster the correlation between standardized rCBV and histologic benchmarks, particularly across a large cohort analysis. Methodology for automatic segmentation of NAWM has recently been reported, demonstrating improvement over manually defined NAWM in the application of tumor grading.34 Future studies would be helpful for comparing such methodology with standardization of rCBV, particularly for differentiating tumor from PTRE in the posttreatment setting. It will also be important to evaluate standardization and normalization methods with evolving consensus recommendations for DSC acquisition, such as with low-flip angle techniques.35
In this study, we spatially matched regional rCBV metrics with regional histologic tumor content, measured in a set of image-localized biopsies from a cohort of patients with recurrent high-grade gliomas. Our results demonstrate that histologic tumor content, at the level of image-localized biopsies, can be predicted by fundamental rCBV metrics such as mean and median rCBV, as well as the FTB metric that reflects the local percentage of voxels with rCBV above/below predetermined thresholds.5,11 These thresholds have previously undergone rigorous histopathologic validation,5,11 and the FTB metric itself has been shown to correlate strongly with global tumor content as a predictor of overall survival,5,11 which can also help to inform clinical decision-making.36 Most interesting, all of the rCBV metrics performed comparably well in distinguishing high-versus-low histologic tumor content when using binary cutoffs (eg, ≥50% tumor, ≥80% histologic tumor content), though the FTB metric appeared to outperform mean and median rCBV when predicting histologic tumor content as a continuous variable (ie, 0%–100% histologic tumor content) (Table 2).
The underlying reasons for this have been suggested previously but likely relate to the microvascular heterogeneity and broad rCBV variability that can be exhibited by GBM, even among tumor subregions.5 These features would make the calculation of mean rCBV susceptible to biases from extremely high or low values, even in a relatively small population of tumor voxels, which could negatively impact the correlation between rCBV magnitude (ie, mean, median rCBV) and variations in histologic tumor proportion. Meanwhile, rCBV magnitude would have less impact on calculation of FTB, except to classify voxels according to minimum threshold requirements. This approach would more purely approximate the volume of histologic tumor content by mitigating influences from varying degrees of tumoral angiogenesis. As an example, a voxel with an rCBV of 1.2 will be classified as tumor in the same manner as another voxel with an rCBV of 12.0. Overall, the use of rCBV, and FTB in particular, offers potential future applications for reducing tissue-sampling errors during surgical biopsy and/or resection of recurrent high-grade gliomas, as well as helping to recover adequate tumor content when molecular profiling is desired.9 As an aside, our cohort did not include patients treated with antiangiogenic therapy such as bevacizumab. Bevacizumab has been shown to decrease both microvessel volume and rCBV values, through mechanisms such as pericyte contraction.37⇓-39 For the aforementioned reasons, thresholding metrics such as FTB would likely be relatively less affected by antiangiogenic-induced rCBV alterations, compared with metrics such as mean rCBV, though further studies are likely needed to fully understand the impact on histologic quantification.
With predictive performance aside, the standardization of rCBV offers methodologic advantages that would help to promote clinical adoption and optimize workflow. There have been recent mounting efforts to homogenize DSC-MR imaging acquisition and analysis toward consensus methodology. The current variability in DSC-MR imaging methodology, from one institution to another, has been implicated as a major reason for the wide variability in reported thresholds that guide clinical diagnosis.10,16,17 The user-based selection of normal brain structures (for rCBV normalization) represents a fundamental process of rCBV map generation that can impact the variability of rCBV values and thresholds, not only across different institutions but at the same institution and within the same patient across repeat examinations.5,20,23
In our study, the intraclass correlations remained high when comparing 2 users defining NAWM regions. In reality, however, this finding may underestimate interrater reliability because we use a highly specific system of NAWM ROI placement in our practice. It would likely be difficult to replicate this level of agreement across different institutions and in routine practice without investing in expert user training to maintain consistency for this postprocessing step. The adoption of rCBV standardization would eliminate the need for user-based input for normalization, which represents an important step toward consensus methodology. One of the key advantages of standardization is that it can be applied to rCBV maps collected at any field strength and for any vendor. This advantage is supported by the positive results obtained with standardized rCBV maps in several clinical trials (American College of Radiology Imaging Network 6677, American College of Radiology Imaging Network 6684) that were collected at both 1.5T and 3T in a range of scanner vendors and at many different institutions.40,41 As explained in the original article on the standardization method,32 standardization calibration files are created for a given protocol. Standardization is therefore robust against slight variations in TE and TR, for example. Furthermore, the fully automated nature of rCBV standardization will also help streamline clinical workflow. By eliminating the need for expert user training, staffing of these user experts, and supervisor oversight (all currently needed for rCBV normalization), the adoption of rCBV standardization (over rCBV normalization) would address potential existing obstacles to integrating DSC-MR imaging in many clinical practices.
We recognize potential limitations to our study. First, image distortions and brain shift following craniotomy could lead to misregistration errors. To compensate, neurosurgeons used small craniotomy sizes to minimize brain shift and also visually validated stereotactic image location with intracranial neuroanatomic landmarks to help correct for random brain shifts. We also used rigid-body coregistration of stereotactic and advanced MR imaging to help reduce possible geometric distortions.29,42 Overall, our experience suggests combined misregistration at approximately 1–2 mm from both brain shift and registration techniques, which is similar to that from previous studies using stereotactic needle biopsy.43 We also acquired DSC-MR imaging data from different MR imaging vendors at 2 different clinical practices. We used identical DSC-MR imaging acquisition parameters but recognize that subtle variability in DSC data could persist. Nonetheless, we also think that this variability could be reflective of the real-world variability that could be encountered in clinical practice and potentially lends strength to the study, given the relatively robust results.
CONCLUSIONS
The standardization of rCBV achieves similar diagnostic accuracy compared with normalized rCBV in diagnosing and quantifying recurrent high-grade glioma tumor burden relative to nontumoral posttreatment radiation effects following standard multimodal therapy. Compared with normalized rCBV, the standardization of rCBV would also eliminate the need for user-based input for normalization (and resulting variabilities in measurement), which represents an important step toward workflow optimization and consensus methodology.
Footnotes
This work was supported by grants NS082609 (NIH), CA221938 (NIH), CA220378 (NIH), CA158079 (NIH), Mayo Clinic Foundation, James S. McDonnell Foundation, Ben and Catherine Ivy Foundation, and the Arizona Biomedical Research Commission (ABRC).
Disclosures: Kris A. Smith—UNRELATED: Payment for Lectures Including Service on Speakers Bureaus: Metabolic Health; Patents (Planned, Pending or Issued): OsteoMed. Richard S. Zimmerman—UNRELATED: Consultancy: NeuroPage, Medtronic. Leslie C. Baxter—RELATED: Grant: National Cancer Institute, Comments: My institution received funding as part of the grant under which this work was performed*; UNRELATED: Employment: Barrow Neurological Institute. Kristin R. Swanson—RELATED: Grant: NS082609, CA221938, CA220378, Mayo Clinic Foundation, James S. McDonnell Foundation, Ivy Foundation, Arizona Biomedical Research Commission*; UNRELATED: Board Membership: James S McDonnell Foundation Scientific Board; Payment for Development of Educational Presentations: multiple patents and applications but no funds. Christopher C. Quarles—RELATED: Grant: National Institutes of Health.* Kathleen M. Schmainda—RELATED: Grant: National Institutes of Health*; UNRELATED: Stock/Stock Options: IQ-AI Ltd, Comments: ownership interest; Other: Imaging Biometrics LLC, Comments: spouse’s salary. Leland S. Hu—RELATED: Grant: U01 CA220378, R01 CA221938, R21 NS082609, Mayo Clinic Foundation, Arizona Biomedical Research Commission*; UNRELATED: Patents (Planned, Pending or Issued): multiple patents but no money paid. *Money paid to the institution.
Indicates open access to non-subscribers at www.ajnr.org
References
- Received September 30, 2019.
- Accepted after revision December 23, 2019.
- © 2020 by American Journal of Neuroradiology