Abstract
BACKGROUND AND PURPOSE: Recently identified molecular subgroups of medulloblastoma have shown potential for improved risk stratification. We hypothesized that distinct MR imaging features can predict these subgroups.
MATERIALS AND METHODS: All patients with a diagnosis of medulloblastoma at one institution, with both pretherapy MR imaging and surgical tissue, served as the discovery cohort (n = 47). MR imaging features were assessed by 3 blinded neuroradiologists. NanoString-based assay of tumor tissues was conducted to classify the tumors into the 4 established molecular subgroups (wingless, sonic hedgehog, group 3, and group 4). A second pediatric medulloblastoma cohort (n = 52) from an independent institution was used for validation of the MR imaging features predictive of the molecular subtypes.
RESULTS: Logistic regression analysis within the discovery cohort revealed tumor location (P < .001) and enhancement pattern (P = .001) to be significant predictors of medulloblastoma subgroups. Stereospecific computational analyses confirmed that group 3 and 4 tumors predominated within the midline fourth ventricle (100%, P = .007), wingless tumors were localized to the cerebellar peduncle/cerebellopontine angle cistern with a positive predictive value of 100% (95% CI, 30%–100%), and sonic hedgehog tumors arose in the cerebellar hemispheres with a positive predictive value of 100% (95% CI, 59%–100%). Midline group 4 tumors presented with minimal/no enhancement with a positive predictive value of 91% (95% CI, 59%–98%). When we used the MR imaging feature–based regression model, 66% of medulloblastomas were correctly predicted in the discovery cohort, and 65%, in the validation cohort.
CONCLUSIONS: Tumor location and enhancement pattern were predictive of molecular subgroups of pediatric medulloblastoma and may potentially serve as a surrogate for genomic testing.
ABBREVIATIONS:
- CP/CPA
- cerebellar peduncle/cerebellopontine angle cistern
- FSL
- fMRI of the Brain Software Library
- SHH
- sonic hedgehog
- WNT
- wingless
Medulloblastoma is the most common malignant pediatric brain tumor, accounting for 40% of childhood tumors in the posterior fossa.1 Genomic characterization of medulloblastoma has recently demonstrated that medulloblastomas can be subdivided into 4 distinct molecular subgroups: wingless (WNT), sonic hedgehog (SHH), group 3, and group 4.2⇓–4 These subgroups have shown different clinical behaviors and may benefit from subgroup-specific treatments. Despite the potential clinical utility of genomic analyses, their translation into clinical practice to improve treatment outcomes in children can be hampered by cost or lack of access to molecular-analysis tools when treatment is initiated. Immunohistochemistry markers have shown utility, but their use is still not widespread and interpretation can be challenging.2,5
MR imaging, on the other hand, is performed in all patients with brain tumor and remains the primary method for diagnosis, surgical guidance, and surveillance of these tumors. Therefore, MR imaging features specific to molecular subgroups of medulloblastoma could facilitate the real-time translation and integration of genomic-based studies into clinical practice. Prior studies have shown that medulloblastomas present with heterogeneous imaging features, including location and enhancement patterns.6 These phenotypic radiologic features may reflect underlying differences in tumor biology.7,8 In this study, we hypothesized that distinct MR imaging features predict molecular subgroups of pediatric medulloblastoma.
Materials and Methods
Patient Cohorts
After institutional review board approval, we retrospectively identified a cohort of patients with medulloblastoma from January 1998 to January 2013 at Lucile Packard Children's Hospital (Stanford University, Palo Alto, California). Patients with both treatment-naive MR imaging and surgical tissue available for molecular analysis were included in the discovery cohort. An independent validation cohort of children with the same inclusion criteria was assembled from the Hospital for Sick Children (Toronto, Ontario, Canada).
Molecular Analysis
NanoString-based assay (http://www.nanostring.com) was performed to classify the medulloblastoma into the 4 main molecular subgroups (WNT, SHH, group 3, and group 4) on the basis of gene-expression profiling, as previously described.9 For most of the patient cohort, molecular analysis was conducted on formalin-fixed paraffin-embedded tissue obtained at diagnosis. One patient underwent molecular subgrouping based on frozen tissue.
Imaging Technique
All patients from the discovery cohort underwent brain MR imaging at 1.5 or 3T (Signa or Discovery 750; GE Healthcare, Milwaukee, Wisconsin). We obtained the following sequences: axial and coronal T2 FSE (TR/TE, 2700/100 ms), axial FLAIR (TR/TE, 9000/120 ms; TI, 2200 ms), precontrast T1 spin-echo and contrast-enhanced T1 spoiled gradient-recalled echo (TR/TE, 8/3 ms; 1-mm section thickness, 0 skip), followed by 2 planes of contrast-enhanced T1 spin-echo (TR/TE, 600–700/20 ms; 5-mm section thickness, 0.5 skip). Many, but not all, patients underwent DWI (TR/TE, 8300/70 ms at 1.5T and 10,000/80 ms at 3T; b-value of 1000 s/mm2; 3 directions; 4-mm thickness, 0 skip) and 2D gradient recalled-echo imaging (TR/TE, 570/30 ms at 1.5T; 700/25 ms at 3T). For the validation cohort, pre- and at least 2-plane postcontrast T1WI obtained at either 1.5T or 3T was used.
Imaging Analysis
Two board-certified radiologists (K.W.Y., J.A. [second-year neuroradiology fellow]) independently reviewed the MR images of the discovery cohort blinded to clinical, pathologic, and molecular data. Consensus for discordant readings was decided between the 2 attending neuroradiologists with Certificates of Added Qualification (K.W.Y. [7 years' experience] and P.B. [>30 years' experience]).
The MR imaging features assessed included the following: tumor location, enhancement pattern, cysts/cavities, hemorrhage/mineralization, intracranial or leptomeningeal seeding, tumor margin, necrosis as suggested by ring-enhancement, and region-of-interest-based ADC analysis, as previously described.6 Specifically, “tumor location” was defined as midline vermian/fourth ventricle, cerebellar hemisphere, or cerebellar peduncle/cerebellopontine angle cistern (CP/CPA). “Tumor margin” was characterized as ill-defined if >50% of the margin could not be distinguished from the surrounding cerebellar parenchyma on the basis of all imaging sequences. “Enhancement pattern” was defined as minimal/none if <10% was estimated to enhance, solid if >90% of the tumor volume was estimated to enhance, and heterogeneous if varying degrees of enhancement were seen in 10%–90% of the tumor volume on the basis of radiologists' visual assessments. Low signal on 2D gradient recalled-echo was used to detect hemorrhage/mineralization.
For the validation cohort, only the MR imaging features found to be significant by the discovery cohort were used for tumor characterization. Two reviewers (S. Perreault and K.W.Y.) independently performed the imaging analysis blinded to clinical, histologic, and molecular information. Any discrepant reads were further weighed in by a third neuroradiologist (P.B.) for a final consensus read.
Stereospecific Computational Map (“Location Heat Maps”)
ROIs outlining the tumor margin were manually drawn in every axial section by using OsiriX Imaging Software (http://www.osirix-viewer.com [A.S.A.]), and proper placement was confirmed by a neuroradiologist (K.W.Y.). All images for each patient were registered to a 1.0-mm isotropic brain atlas (Montreal Neurological Institute) by using a mutual-information algorithm and a transformation algorithm in 3D Slicer (www.slicer.org), followed by visual inspection and a consensus by 2 independent raters (A.S.A., T.T.L.) to ensure optimal alignment.10 All lateralized medulloblastomas were projected to one side for analysis. After image registration, the resulting transformation matrix was used to map the ROI coordinates to the Montreal Neurological Institute atlas space followed by a second round of visual inspection and consensus by the raters. The frequency of tumor occurrence in each voxel of the Montreal Neurological Institute atlas space was then calculated to create a probabilistic radiographic atlas visualized as 3D heat maps in Slicer and by using FSLView (http://fsl.fmrib.ox.ac.uk/fsl/fslview/).
Neurosurgical Evaluation
Operative reports were independently reviewed by a blinded pediatric neurosurgeon (K.C.). Surgical examinations regarding primary tumor locations and areas of brain invasion were recorded and classified as midline vermian/fourth ventricle, cerebellar hemisphere, or CP/CPA.
Pathologic Evaluation
Hematoxylin-eosin–stained slides of formalin-fixed paraffin-embedded material were analyzed by an independent neuropathologist (H.V.) blinded to radiologic, clinical, or molecular information. Medulloblastomas were categorized according to the 2007 WHO Classification of Tumors of the Central Nervous System.11
Statistical Analysis
Statistical analyses were performed by using the Fisher exact test and χ2 analysis for categoric data. A multivariable logistic regression model was developed to identify significant predictors of the medulloblastoma subgroup. We explored potential multicolinearity among the independent variables, examining changes in significance and exploring significant associations among independent variables. Pseudo R-squared goodness of fit ascertained by using the Cox and Snell method (Statistical Package for the Social Sciences Statistics, Version 20.0; IBM, Armonk, New York) was used for the above analyses. NanoString prediction and normalization of data were performed by using the R statistical environment (Version 2.5.1, http://www.r-project.org) as previously described.9
Statistical analysis to identify areas of differential involvement consisted of first constructing a contingency table comparing 2 differential phenotypes (eg, WNT versus groups 3 and 4, or SHH versus groups 3 and 4) and the presence of tumor versus no tumor involvement for each image voxel, with a 2-tailed Fisher exact test performed on a voxelwise basis (using the FSL tool Randomize [http://www.fmrib.ox.ac.uk/fsl/randomize/]). The resulting P value representing the contingency table (ie, tumor involvement of voxels in a particular phenotype) had <5% probability of occurring by chance. Permutations with the threshold-free cluster enhancement method previously described were applied to correct for multiple comparisons, and a family-wise error rate, to ensure a false discovery rate of <0.05.12 A total of 3876 unique permutations were performed for an exhaustive test for WNT versus groups 3 and 4, and 10,000 permutations, for SHH versus groups 3 and 4. The resulting corrected P value representing tumor involvement of certain voxels in a particular phenotype had <5% false discovery rate, adjusted for multiple comparisons. These significant voxels were visualized by using FSLView.
Results
Histology, Tumor Staging, and Molecular Subgroups
Forty-seven patients met the inclusion criteria and were included in the discovery cohort (Table 1). Median age at diagnosis was 8.2 years (range, 0.9–33.9 years). The 47 medulloblastomas consisted of 31 classic (66%), 10 large-cell/anaplastic (21%), 4 desmoplastic (9%), and 2 other medulloblastoma histologic features (4%).
Cohort demographics
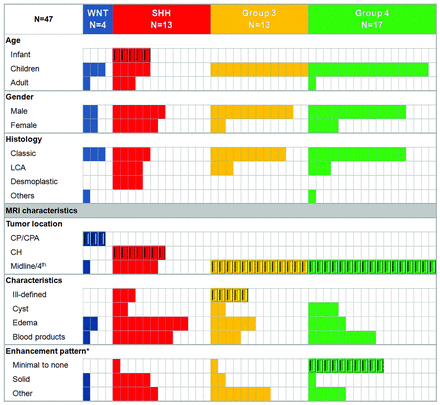
NanoString assay demonstrated 4 WNT (9%), 13 SHH (28%), 13 group 3 (28%), and 17 group 4 (36%) medulloblastoma molecular subgroups (Table 1 and Fig 1).
Patient characteristics of the discovery cohort according to the molecular subgroups and MR imaging features. CH indicates cerebellar hemisphere; LCA, large-cell anaplastic; enhancement pattern (asterisk), minimal to none, <10% tumor volume; solid, >90% tumor volume. Beveled rectangles represent statistical significance (Fisher exact test [P < .005], except for Ill-defined margins [P = .03]).
MR Imaging Correlates of Molecular Features in the Discovery Cohort
Tumor location was highly predictive of the molecular subgroups (Figs 1 and 2 and On-line Figs 1 and 2). Seventy-five percent of WNT tumors occurred along the CP/CPA. This location was unique to this molecular subgroup (P < .001) and was associated with a positive predictive value of 100% (95% CI, 30%–100%). Cerebellar hemispheric location was characteristic of the SHH tumors and accounted for 54% of this molecular subgroup (P ≤ .001, positive predictive value of 100% [95% CI, 59%–100%]). Groups 3 and 4 were primarily midline and occupied the fourth ventricle (100%, P < .001). Tumor location did not significantly differ on the basis of age.
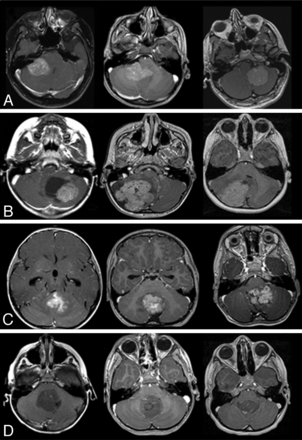
Characteristic MR imaging features according to medulloblastoma molecular subgroups. A, In the top row, characteristic location of WNT tumors in the CP/CPA region is shown. B, In the second row, SHH tumors are predominantly located in the cerebellar hemispheres. C, In the third row, group 3 tumors are located in the midline/fourth ventricle and show enhancement and ill-defined features against the adjacent brain parenchyma. D, In the fourth row, group 4 tumors are also located in the midline fourth ventricle but tend to show minimal or no enhancement.
Group 3 was characterized by an ill-defined tumor margin (63%), a feature not present in other subgroups (10%) (P = .03). Only 3 other tumors demonstrated ill-defined margins, all in the SHH subgroup. Minimal or no enhancement was characteristic of group 4 medulloblastoma, present in 10 patients (59%) compared with only 2 (7%) for the other subgroups (P < .001, positive predictive value of 83% [95% CI, 52%–97%]). Only 1 nonenhancing medulloblastoma belonged to group 3. This feature also distinguished group 4 from group 3 medulloblastomas with a positive predictive value of 91% (95% CI, 59%–98%).
The presence of hemorrhage/mineralization was assessed in 33 (70%) patients who had available 2D gradient recalled-echo sequences but was not found to correlate with specific molecular groups. The other MR imaging features (cysts, peritumoral edema, and tumoral necrosis) were not characteristic of specific molecular subgroups. Twenty-two (47%) patients had available DWI in the discovery cohort (WNT, 2/4; SHH, 8/13; group 3, 5/13; group 4, 7/17). Mean ADC did not significantly differ among the molecular subgroups: WNT, 740 × 10−6 mm2/s (range, 684–796 mm2/s); SHH, 714 × 10−6 mm2/s (range, 655-947 mm2/s); group 3, 733 × 10−6 mm2/s (range, 650–847 mm2/s); group 4, 767 × 10−6 mm2/s (range, 661–813 mm2/s).
Stereospecific Computational Map by Molecular Subgroup
Stereospecific computational analyses stratified by molecular subgroup identified group 3 and 4 medulloblastomas occurring predominantly in the midline/fourth ventricle (significant voxels, P = .007–.05; displayed as red in On-line Figs 1 and 2, third column), compared with WNT localization to the CP/CPA (significant voxels, P = .003–.05; displayed as green in On-line Fig 1, third column) and SHH tumors in the cerebellar hemispheres (significant voxels, P = .02–.05 after multiple-comparisons correction; blue in On-line Fig 2, third column).
Neurosurgical Findings
Surgical inspection regarding the primary tumor location was 100% concordant with the presurgical MR imaging assessment. The WNT tumors arose in the CP/CPA (75%); cerebellar SHH tumors occurred in the cerebellar hemispheres (23% versus 0% [P = .02]), and groups 3 and 4 commonly occurred in the midline/fourth ventricle compared with other subgroups (80% versus 40% [P = .02]). Tumor invasion of the brain stem floor/cerebellar peduncle differed among the groups: Adjacent brain invasion occurred in only 1 case of SHH tumor (9%), whereas invasion occurred in 77% for the other subgroups (P = .001).
A Model for Determining Tumor Molecular Subgroup Using the Discovery Cohort
Multivariable logistic regression showed that location (CP/CPA, cerebellar hemisphere, and midline/fourth ventricle, [P < .001]), pattern of enhancement (P = .001), and definition of tumor margin (P = .01) were predictors of medulloblastoma subgroups. With the logistic regression model based on location, pattern of enhancement, and tumor margins, 69% of tumors were appropriately classified. The multivariable model demonstrated a goodness of fit as assessed by the pseudo R-squared goodness of fit (Cox and Snell method) of 0.76.
Validation Cohort
There was no significant difference in terms of demographics, histology, and molecular subgroup proportions between the discovery (Stanford Lucile Packard Children's Hospital, n = 47 patients) and the validation (Toronto Hospital for Sick Children, n = 52 patients) cohorts (P ≥ .05) (Table 1). T1WI pre- and postgadolinium images were used to address the significant imaging features of tumor location, margin, and enhancement pattern identified by the discovery cohort.
The only predictor from the discovery cohort that was not validated by the validation cohort was the tumor margin, with no significant difference between group 3 and other subgroups (P = .7). This variable was then removed from the multivariate model, and predictors, including location and pattern of enhancement, were used to develop a second model. This second multivariable model demonstrated a goodness of fit as assessed by the pseudo R-squared goodness of fit (Cox and Snell method) of 0.67 in the discovery cohort and 0.63 for the validation cohort. When we applied this model, 66% of medulloblastomas were correctly predicted in the discovery cohort, and 65%, in the validation cohort (Table 2 and On-line Fig 3).
Percentage correct with the model when applied to the discovery cohort and validation cohort
Discussion
Several studies have described heterogeneity of medulloblastomas and have related these features to histologic subgroups, age, or prognosis.6,8 Recently, genomic studies have identified 4 unique molecular subgroups of medulloblastomas (WNT, SHH, group 3, group 4) that are more predictive of clinical behavior and outcome than either tumor histology (classic or “variants” [desmoplastic, large cell/anaplastic, extensive nodularity]) or clinical staging system.2⇓–4 For example, WNT tumors have shown good prognosis regardless of histology, whereas group 3 tumors have worse survival, independent of metastatic stage.2,13
The benefits of translating this information into the clinical arena are numerous, given the significant neurotoxic effects of current therapy.14,15 For example, the low-risk molecular WNT medulloblastoma group could be stratified to surgery and chemotherapy only, without radiation that poses significant risk for cognitive impairment in children, while high-risk medulloblastoma molecular groups (groups 3 and 4) could require multimodal therapies and more frequent tumor monitoring; they would be ideal candidates for new targeted experimental therapies in clinical trials. As noted by Robinson16 in a commentary letter, while no single tumor feature should be used alone to determine tumor subtype or tailor treatment, MR imaging can offer additional opportunities. Because MR imaging is already universally used in brain tumor diagnosis, identifying MR imaging correlates of molecular subgroups could further assist in this endeavor and play a key role when access to molecular analysis is limited.
This is the largest comprehensive study to investigate MR imaging correlates of molecular subgroups of medulloblastoma. Tumor location and pattern of gadolinium enhancement were found to be predictive; by using a regression model, most medulloblastoma molecular subgroups were accurately identified. Furthermore, our model was validated by an independent cohort from a second institution.
Using the NanoString assay, we were able to classify our medulloblastoma cohort into 4 recently established molecular subgroups.2,4 Overall, our cohort recapitulated previously described distinct clinical features of each molecular group.2,4 We observed that SHH tumors accounted for more infants, most adults, and females. Group 3 most commonly presented with metastases at diagnosis; and desmoplastic tumors exclusively belonged in the SHH subgroup.17
According to our model, location was a key feature predictive of molecular subgroup. Our MR imaging–based tumor location was also confirmed by surgical examination. Most WNT tumors arose in the CP/CPA, SHH most commonly involved the cerebellar hemispheres, and groups 3 and 4 were midline, filling the fourth ventricle.
Prior studies have shown that desmoplastic medulloblastomas frequently showed SHH molecular features and tended to involve the cerebellar hemispheres, especially in adults,4,18,19 though figures have ranged from 17% to 100%, depending on the clinical inclusion criteria used in these various studies.8,18⇓⇓⇓–22 A cerebellar hemispheric location of SHH medulloblastomas is consistent with the results of the mouse models that have shown SHH tumor origin from committed granule neuron precursors of the cerebellum.23,24 While studies have also suggested that SHH tumors could arise from the cochlear nucleus in the brain stem in younger patients,25 we did not observe a difference in SHH tumor location between infants and older patients. Our results are also consistent with a study by Teo et al,26 which reported cerebellar hemispheric involvement in 9 of 17 (53%) SHH medulloblastomas regardless of age at diagnosis. In our study, SHH rarely invaded the brain stem, consistent with a prior study that reported brain stem infiltration by WNT but not SHH tumors.22
However, our results of WNT tumors differ from those of Teo et al26 in that while midline location was described for all 5/5 WNT tumors in that study, most of the WNT tumors in our study involved the CP/CPA (8/14). In another study, Gibson et al22 reported 6/6 WNT tumors to be midline but infiltrating the dorsal brain stem. Discrepancies among these studies could be attributed to the small sample sizes. Alternatively, tumors that have cerebellar peduncle origin or involvement may have a “midline” appearance, particularly if they are protruding medially. Previously, Jaiwal et al27 described CP/CPA tumor location in 14 of 140 (10%) medulloblastomas. While molecular analysis was not performed in that study, it would be interesting to test these tumors for the WNT pathway because this figure approximates overall prevalence of WNT tumors.2 While we acknowledge that some WNT tumors are midline, we report that a significant percentage of these tumors involve the CP/CPA, which is a unique feature for this subtype.
A significant percentage of medulloblastomas present minimal or no enhancement.6,8 Rare cases of hemispheric desmoplastic medulloblastoma have shown this feature as well.18 In our study, nonenhancing tumors in the midline/fourth ventricle location were characteristic of group 4 medulloblastomas. This is an important observation because groups 3 and 4 are currently not well-differentiated by using immunohistochemistry markers.2,5 The reason for the lack of enhancement in a significant subset of group 4 tumors remains unknown, but molecular changes associated with vascular permeability might be involved.
We did not find a correlation between molecular subgroups and other MR imaging features in our discovery cohort. A prior study showed that ring enhancement/necrosis and higher mean ADC were more characteristic of large-cell/anaplastic medulloblastoma than classic medulloblastoma.6 This finding could be explained by the fact that large-cell/anaplastic histology can be present in 3 different molecular subgroups (group 3, group 4, and SHH),2,17 or even in WNT tumors.13 Tumor margin against the brain parenchyma was a useful predictor in our discovery but not in the validation cohort. However, evaluation of tumor margins was significantly limited because only pre- and postcontrast T1WI were uniformly available in our validation cohort, without a complete MR imaging dataset inclusive of multiplanar T2WI (axial and coronal), FLAIR, and thin-section presurgical or spoiled gradient-recalled echo sequences in the discovery cohort. The definition of margins could have been either under- or overestimated, thereby potentially limiting its usefulness. Because this feature could not be used in our validation cohort, the first model could not be tested. A new model from our discovery cohort without this limiting feature was developed. Our final algorithm was, therefore, influenced by our discovery cohort but only because one of the features could not be tested.
This study was also limited by its retrospective nature. Some heterogeneity in imaging protocols reduced the sample size for some components of our analysis. For example, features such as mean ADC and hemorrhage/mineralization did not show a significant difference between subgroups in our exploratory cohort, but type II errors remain possible. To overcome such limitations, our plan includes a multicenter prospective study.
Conclusions
MR imaging features of tumor location and enhancement pattern were correlated with specific molecular subgroups of medulloblastoma and were validated by an independent cohort. This study represents an important step in using MR imaging as a surrogate to predict molecular subgroups of medulloblastoma. Future study that incorporates quantitative MR imaging signatures including perfusion, MR spectroscopy, high-order diffusion, and susceptibility metrics, could add insight into formulating a more robust radiogenomics model for medulloblastoma.
Acknowledgments
We thank Drs Patrick D. Barnes, Jalal B. Andre, and Lex A. Mitchell for their work on tumor imaging annotations and DICOM data extraction, Amarjeet Grenwall and Dory Palacio for their help in tissue processing, and Dr Patryk Skowron for his involvement in NanoString-based assays.
Footnotes
Yoon-Jae Cho and Kristen W. Yeom are co-senior authors.
Disclosures: Sébastien Perreault—RELATED: fellowship grants from Fonds de la Recherche en Santé du Québec and Irma-Levasseur. Vijay Ramaswamy—RELATED: clinical fellowships from the Canadian Institutes of Health and Alberta Innovates Health Solutions, providing salary support for work both related and unrelated to the current work. He also discloses the Canadian Cancer Society Research Institute travel award, to attend the International Society of Paediatric Oncology 2013 meeting in Hong Kong. Eric Bouffet—RELATED: grant support from the Medulloblastoma Advanced Genomics International Consortium (Principal Investigator: Dr Michael Taylor).* Paul Fisher—UNRELATED: payment for teaching lectures in general neurology for the American Academy of Pediatrics. He is also an associate editor on the Journal of Pediatrics, a paid position with Elsevier. Yoon-Jae Cho—UNRELATED: honoraria for grand rounds lectures at St. Jude, Memorial Sloan Kettering Cancer Center, and Texas Children's. *Money paid to the institution.
Sébastien Perreault was a Beverly and Bernard Wolfe Pediatric Neuro-Oncology fellow at Lucile Packard Children's Hospital at Stanford University.
This work was supported by Fonds de Recherche en Santé du Québec (S. Perreault), Justine Lacoste Foundation (S. Perreault), St. Baldrick's Foundation (Y.J. Cho), and clinical fellowships from the Canadian Institutes of Health Research and Albert Innovates Health Solutions (V. Ramaswamy).
Previously presented in abstract form at: Annual Meeting of the Society for Neuro-Oncology, November 21–14, 2013; San Francisco, California.
Indicates open access to non-subscribers at www.ajnr.org
References
- Received February 17, 2014.
- Accepted after revision March 20, 2014.
- © 2014 by American Journal of Neuroradiology