Abstract
BACKGROUND AND PURPOSE: Both IDH1 mutation and MGMT promoter methylation are associated with longer survival. We investigated the ability of imaging correlates to serve as noninvasive biomarkers for these molecularly defined GBM subtypes.
MATERIALS AND METHODS: MR imaging from 202 patients with GBM was retrospectively assessed for nonenhancing tumor and edema among other imaging features. IDH1 mutational and MGMT promoter methylation status were determined by DNA sequencing and methylation-specific PCR, respectively. Overall survival was determined by using a multivariate Cox model and the Kaplan-Meier method with a log rank test. A logistic regression model followed by ROC analysis was used to classify the IDH1 mutation and methylation status by using imaging features.
RESULTS: MGMT promoter methylation and IDH1 mutation were associated with longer median survival. Edema levels stratified survival for methylated but not unmethylated tumors. Median survival for methylated tumors with little/no edema was 2476 days (95% CI, 795), compared with 586 days (95% CI, 507–654) for unmethylated tumors or tumors with edema. All IDH1 mutant tumors were nCET positive, and most (11/14, 79%) were located in the frontal lobe. Imaging features including larger tumor size and nCET could be used to determine IDH1 mutational status with 97.5% accuracy, but poorly predicted MGMT promoter methylation.
CONCLUSIONS: Imaging features are potentially predictive of IDH1 mutational status but were poorly correlated with MGMT promoter methylation. Edema stratifies survival in MGMT promoter methylated but not in unmethylated tumors; patients with methylated tumors with little or no edema have particularly long survival.
ABBREVIATIONS:
- CI
- confidence interval
- GBM
- glioblastoma multiforme
- IDH1
- isocitrate dehydrogenase-1
- IQR
- interquartile range
- MGMT
- O-6-methylguanine-DNA-methyltransferase
- OR
- odds ratio
- nCET
- non-contrast enhancing tumor
- PCR
- polymerase chain reaction
- ROC
- receiver operator curve
GBMs are the most aggressive and lethal primary brain tumors.1 Standard therapy for GBM is maximal tumor resection with radiation therapy and temozolomide treatment. This is commonly followed by antiangiogenic therapy with bevacizumab at recurrence.2 GBM can arise de novo or from degeneration of lower grade tumors (secondary GBM).3 Mutations in the citric acid cycle enzyme IDH1 have recently been implicated in gliomagenesis and are found in approximately 70%–80% of secondary glioblastomas but are much more rare (<10%) in primary GBMs.4⇓⇓⇓–8 IDH1 mutations are associated with a distinct gene expression profile9 (in particular, the proneural subset of malignant gliomas) and are considered an independent prognostic indicator in these patients.9,10 Epigenetic silencing of the DNA repair enzyme MGMT is another molecular feature of GBM that has both prognostic and predictive significance because methylation is associated with better outcomes as well as response to temozolomide therapy.11
In addition to molecular signatures, several MR imaging−derived features of GBM also correlate with length of survival. For instance, multiple studies have shown that edema and necrosis are associated with poor outcomes.12⇓–14 Other imaging features have been shown to be potentially prognostic in patients with gliomas, including cysts,15 enhancement,12,16,17 multifocality,12 and location.18,19 Some of these imaging features, such as multifocality,20 enhancement,21 location,18 and edema,22 have known molecular correlates.23 We hypothesized that some of these imaging features reflect differences in molecular signatures such as mutation of IDH1 or MGMT promoter methylation. Therefore, we analyzed the ability of these potentially prognostic imaging features derived from standard MR imaging sequences to predict IDH1 mutational status and MGMT promoter methylation, to develop noninvasive easily acquired biomarkers of these important molecular subtypes of GBM.
Materials and Methods
Patients
All patients participating in this retrospective study signed institutional review board−approved informed consent, agreeing to participation in a study of imaging analysis and clinical outcomes. Outcome data from some subsets of these patients have been previously published.24,25 Data acquisition was performed in compliance with all applicable Health Insurance Portability and Accountability Act regulations. Patients were selected from the neuro-oncology data base of our institution on the basis of the following criteria: 1) pathology-confirmed GBM based on modified World Health Organization grading system, 2) baseline (presurgical) MR imaging scan, 3) age ≥18 years, and 4) treatment including radiation therapy and temozolomide. Of the 202 patients, 96 had gross total resection, 94 had subtotal resection, and 12 had biopsy only (based on standard imaging). Cases included those associated with a previously published study.26 Follow-up scans were obtained at approximately 4- to 6-week intervals. Steroid doses for patients at the time of initial scanning were not available in most cases. The study spanned 1999–2009. At the time of last assessment (May 2010), 140 of the 202 patients (69%) had died. Most patients in the study received bevacizumab as part of their treatment (186/202, 92%). This was administered “up-front,” that is, 3–6 weeks after tumor resection concurrent with radiation therapy and temozolomide, or they received bevacizumab at tumor recurrence.27 Baseline patient data are shown in Table 1, segregated into groups on the basis of bevacizumab treatment.
Baseline characteristics
Imaging Acquisition
MR imaging was performed on 1.5T or 3T scanners and typically included axial T1-weighted (TR, 400 ms; TE, 15 ms; section thickness, 5 mm), T2-weighted fast spin-echo (TR, 4000 ms; TE, 126–130 ms; section thickness, 5 mm), and gadodiamide- (Omniscan; GE Healthcare, Little Chalfont, Buckinghamshire, United Kingdom; 0.1 mmol/kg) or gadopentetate dimeglumine−enhanced (Magnevist; Bayer Healthcare Pharmaceuticals, Montville, New Jersey; 0.1 mmol/kg) axial and coronal T1-weighted images (TR, 400 ms; TE, 15 ms; section thickness, 3 mm), with an FOV of 24 cm and a matrix size of 256 × 256. Postcontrast images were acquired immediately following contrast injection.
Scoring Imaging Features
Tumors were assessed for size, enhancement, nCET, necrosis, edema, cysts, multifocality, contact with ventricles or neocortex, and location based on prior work,12 by a neuroradiologist (W.B.P.) blinded to molecular and clinical data as well as outcomes. Briefly, moderate/severe edema was defined as edema extending >1 cm from the margin of the tumor on T2-weighted images; otherwise edema was scored as little, or if absent, none. nCET was graded as positive or negative (for Kaplan-Meier analysis) and was also qualified as none; minimal; approximately 25%, 50%, 75%; or almost all or all (for the ROC analysis, see below), as judged by the reader (W.B.P.). Nonenhancing tumor was defined as areas of intermediate T2-weighted hyperintensity, less than the intensity of CSF, and corresponding to a region of T1-weighted hypointensity, which was associated with mass effect and architectural distortion, including blurring of the gray-white junction and/or expansion of the deep nuclei, and which showed no obvious enhancement. This method was shown to have high interobserver agreement in a prior study.21 Size was determined on the basis of postcontrast T1-weighted images and was measured in longest dimension in centimeters. Enhancement was scored positive for tumors that demonstrated unequivocal increased signal intensity on T1-weighted images following intravenous contrast administration. “Multifocal tumors” were defined as having >1 area of tumor separated by normal brain signal intensity on T2-weighted images. If a secondary lesion fell within the T2-weighted signal-intensity change of the dominant nodule, then the lesion was classified as a “satellite lesion.” “Necrosis” was defined as a region or regions of peripheral and irregular enhancement surrounding areas of high T2-weighted signal intensity. A “cystic tumor” was defined as having an area with peripheral/rim enhancement measuring >1 cm in diameter, demonstrating a thin uniform wall with central high T2-weighted signal intensity approximating that of CSF. “Location” was defined as centered in the frontal, parietal, temporal, occipital, insula or within the posterior fossa, or confined to the deep nuclei (basal ganglia/thalamus). Large tumors that were not clearly centered in a single lobe were scored by the lobes involved (frontal-temporal, frontal-parietal, and so forth).
Molecular Analysis
MGMT promoter methylation status was available for 190 patients and was determined from formalin-fixed paraffin-embedded tissue samples, as previously described.16 IDH1 mutation was determined by genomic sequencing analysis to identify brain tumor samples containing either wild-type IDH1 or mutations altering amino acid R132. Genomic DNA was isolated from 50–100 mg of brain tumor tissue by using standard methods and a PCR was used to amplify a 295 base pair fragment of the genomic DNA that contains both the intron and second exon sequences of human IDH1, allowing assignation of mutation status assessed by standard molecular biology techniques.
Statistical Methods
Overall survival from the time of tumor resection was recorded, and the median survival with the IQR and the mean patient age with the SD were generated. A test of the proportional hazards assumption was used after fitting a multivariate Cox model, which included MR imaging−derived imaging features, MGMT promoter methylation status, IDH1 mutation status, age, and then the corresponding 95% CIs were generated. The Kaplan-Meier method with the log rank test was used to estimate overall survival on the basis of MGMT promoter methylation status (also stratifying by nCET and edema) and IDH1 mutation status. The level of significance for Kaplan-Meier plots with >2 survival curves represents the overall comparison (based on the log rank test), indicating that at least 1 group is statistically different from the other groups, when P is <.05. A recursive portioning analysis was used to compare survival on the basis of methylation status and edema.
To compare each clinical and imaging feature between 2 groups from the recursive analysis, we performed a t test or Wilcoxon rank sum test for continuous variables, depending on the distribution (normal versus not), and a χ2 test for categoric variables. The Spearman ρ correlation coefficient was calculated to characterize the association between tumor location and the MGMT promoter methylation/IDH1 mutational status. A multiple logistic regression analysis was performed for molecular types by using imaging features as covariates. A backward variable selection was used with a significance threshold of 0.1 to model IDH1 status, and then ROC analysis was performed to evaluate the model by using an area under the curve analysis. Robustness of the model for classifying IDH1 status was confirmed with 20,000 replication bootstrapping, and the bias-corrected 95% CI was reported. For all analyses, a P value of <.05 was accepted as significant. Statistical analysis was performed with STATA (StataCorp, College Station, Texas).
Results
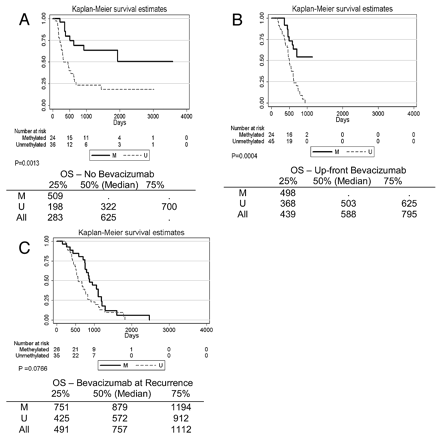
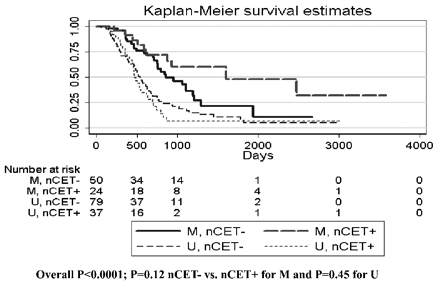
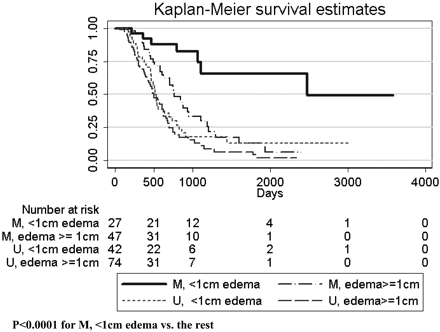
MGMT promoter methylation was found in 74/190 (39%) patients. MGMT promoter methylation status was associated with better survival (P < .0001, log rank test). This relationship was true regardless of whether patients never received bevacizumab (P = .0013) or received bevacizumab up-front (P = .0004). Methylation status also appeared to stratify survival in patients who received bevacizumab at recurrence, though this did not meet statistical significance (P = .077, Fig 1). Adding nCET to methylation status in the Kaplan-Meier analysis appeared to further stratify survival (Fig 2), though the results were not statistically significant in a pair-wise analysis (ie, overall there was a difference in survival curves, but when comparing selected pairs of subgroups, P values were >.05). However, when edema was used to stratify survival, patients with methylated tumors but without edema lived significantly longer than patients with unmethylated tumors or methylated tumors with edema (Fig 3). Using a recursive partitioning analysis for MGMT promoter methylation status, we found that methylated tumors without edema had a median survival of 2476 days (95% CI, 1065 not reached), compared with methylated tumors with edema (762 days; 95% CI, 610–953), unmethylated tumors without edema (552 days; 95% CI, 464–702), and unmethylated tumors with edema (501 days; 95% CI, 412–623).
Methylation status stratifies survival in patients not treated with bevacizumab (A) and in patients treated with bevacizumab concurrent with radiation-temozolomide (B). C, A similar pattern is seen in patients treated with bevacizumab at recurrence, though this did not reach statistical significance.
Kaplan-Meier curves for methylated-versus-unmethylated tumors stratified by nCET. The overall P value represents the overall comparison by log rank test, indicating that at least 1 group is different from the others.
Kaplan-Meier analysis for patients (n = 190) with known methylation status, stratified by edema levels. Note that methylated tumors without significant edema have much longer survival than either unmethylated tumors or methylated tumors with edema.
The difference in survival between methylated tumors without edema and the remainder (2476 versus 586 days, P < .0001) was not due to age or extent of resection, which was not significantly different between the 2 groups. The only difference between the 2 groups (methylated tumors without edema versus the remainder) in imaging features was that the methylated tumors without edema were slightly smaller (unidimensional largest diameter mean was 4.2 cm for methylated tumors with little/no edema versus 5.1 cm for remainder tumors, t test, P = .019). Methylation status was mildly correlated with the presence of multifocal tumors and tumors with satellite lesions (−0.17, P = .023 and 0.18, P = .012, respectively, χ2 test) but did not correlate with any of the other imaging features. Using a multiple logistic regression analysis, we found that imaging features were only a little better than chance (66% accuracy) at predicting methylation status.
Of 202 tumors, 63 (31%) were nCET+. There was an inverse correlation between nCET+ and edema (r = −0.36; P < .0001; OR, 0.34; P < .001) as previously shown.12 Multifocality, which is thought to be associated with poor outcomes,12 was less common for methylated compared with unmethylated tumors (OR, 0.20; P = .038) but was more common among nCET+ compared with nCET− tumors (OR, 7.94; P = .001).
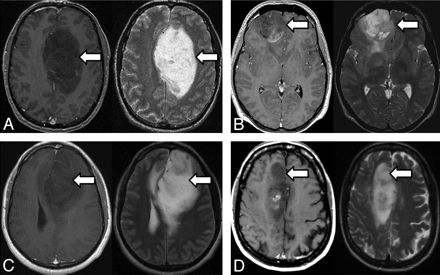
IDH1 mutation was present in 14/202 (6.9%) tumors, and was associated with longer survival (P = .002, log rank test). All IDH1 tumors were nCET+ (Fig 4). Most IDH1 tumors (11/14, 79%) involved the frontal lobe versus 69/188 (37%) wild type tumors, as reported previously for this dataset.26 Thus the OR for frontal lobe involvement for mutant versus wild type was 6.3 (95% CI, 1.7–23.5). The IDH1 mutation was not an independent predictor of survival in a multivariate analysis that included imaging features and methylation status (Table 2). This is likely due to the association of the IDH1 mutation with MGMT promoter methylation (OR for methylated tumors versus unmethylated tumors being IDH1 mutants: 3.07, P = .053; Spearman ρ = 0.15, P = .044). Methylation status was available for all 14 IDH1 mutant tumors. Of these, 9 (64%) were found to be methylated.
A−D, Four examples of IDH1 mutant tumors on MR imaging. For each set of paired images, T1 postcontrast images on the left are shown with the corresponding T2-weighted image on the right. Note abundant nonenhancing tumor (arrows) and frontal lobe location.
Multivariate analysis of molecular markers and imaging features in relationship to survivala
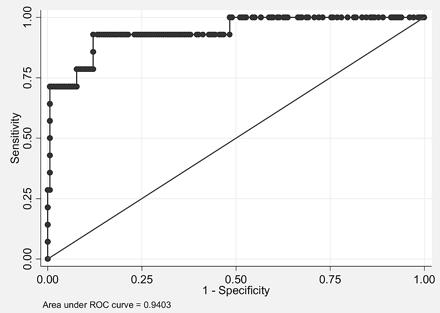
To test whether imaging could predict IDH1 status, we performed a logistic regression analysis by using all imaging features as covariates. A higher percentage of nCET, larger size of tumor, presence of cysts, and presence of satellites all correlated with IDH1 mutant tumors by using a backward variable selection with a threshold of 0.1. The area under the curve for IDH1 mutational status was 0.94 (Fig 5) with bootstrap bias-corrected 95% CI, 0.85–0.99. Accuracy was 97.5% with a sensitivity of 71.4% and a specificity of 99.5% (note that bootstrap bias−corrected 95% CIs were 95.0%–100%, 42.9%–100%, and 97.8%–100% for accuracy, sensitivity, and specificity, respectively).
ROC analysis of imaging features used to predict IDH1 mutational status. Combining percentage nCET, tumor size, cyst, and satellites allows a 94% accuracy in classifying tumors in this test set with 14 IDH1 mutants.
Discussion
Imaging correlates of gene expression may provide important insight into subtypes of GBM. For example, Pope et al21 compared gene expression in completely-versus-incompletely enhancing tumors and found overexpression of genes associated with hypoxia, angiogenesis, and edema in the former. Similarly, Van Meter et al28 and Diehn et al29 examined genes that were differentially expressed on the basis of contrast enhancement and areas of central necrosis, again noting upregulation of genes associated with angiogenesis within areas of contrast enhancement. Barajas et al30 analyzed the relationship between gene expression and radiographic images, including physiologic imaging techniques such as perfusion and diffusion MR imaging, showing that regions of tumor with high blood volume and/or low apparent diffusion coefficient express gene profiles associated with angiogenesis and tumor aggressiveness. In contrast to angiogenic gene expression associated with edema, necrosis, and shortened survival, the IDH1 mutation and MGMT promoter methylation status define molecular subtypes of glioblastoma with a favorable prognosis. We hypothesized that these molecular signatures may be reflected in imaging features derived from standard MR imaging. Therefore, we analyzed the relationship between MR imaging features and these GBM subtypes in a large cohort of patients as a first step in developing noninvasive biomarkers of these groups that may be useful, particularly when tissue-based testing is inconclusive or not possible.
As previously shown,31 both the IDH1 mutation and MGMT promoter methylation were associated with longer survival. MGMT promoter methylation has been associated with susceptibility to temozolomide treatment.11 Temozolomide is 1 of 3 medications approved by the FDA for the treatment of glioblastoma, in addition to carmustine32 and, more recently, the anti-angiogenic agent bevacizumab.2 All the patients in the current study received temozolomide, so the response to this treatment based on methylation status could not be tested. However, we did find that methylation status stratified survival, regardless of bevacizumab therapy.
In a multivariate analysis, we found that MGMT promoter methylation was prognostic of better outcome, but IDH1 status was only significant in a univariate analysis. There are 2 likely reasons why IDH1 status was not independently prognostic in the multivariate model. One is that there was a strong correlation between IDH1 mutation and MGMT promoter methylation as others have previously found.33 Second, the IDH1 mutation is relatively rare in GBM because it is associated with secondary but not primary tumors.7 We found that only 6.9% of tumors in our cohort were IDH1-mutated, similar to the incidence in a previous report.31 Thus the low number of IDH1 mutants and strong correlation with MGMT promoter methylation may explain the lack of independent impact on prognosis in the multivariate analysis.
As for imaging features, edema and multifocality were associated with poor outcome, as previously demonstrated.12,34 Rim enhancement has been reported to be associated with unmethylated primary GBM, whereas a mixed-nodular enhancement pattern was associated with methylated and secondary GBM.23,35,36 We did not find a significant difference in edema levels or tumor location between methylated and unmethylated tumors, similar to findings in prior reports.35,36 However, we did find that edema was able to stratify survival in methylated but not in unmethylated tumors. Thus edema levels may be particularly important for patient prognosis in the MGMT promoter methylated tumor subset.
MGMT promoter methylation is more common in the proneural than the mesenchymal subset of GBM.33 Proneural tumors have a better prognosis than other glioblastoma subtypes.37,38 It may also be that tumors with little or no edema also are more likely to fall within the proneural subset, but this hypothesis has not been formally tested, to our knowledge. Previous reports have shown that MR imaging texture can predict MGMT promoter methylation status with fairly high accuracy (71%).35 We did not perform a texture analysis. We did not find any imaging features that correlated well with MGMT promoter methylation, so it seems that predicting methylation status from MR imaging without postprocessing analyses or advanced imaging techniques may be challenging.
Results from the current study suggest that imaging features could be used to predict IDH1 mutation with 97.5% accuracy. Although this needs to be confirmed in a large prospective trial, these results suggest that simple imaging features might be able to serve as a useful biomarker of IDH1 status. For example, all IDH1 mutant tumors were nCET+, and most IDH1 tumors involved the frontal lobe. Because increased vascular endothelial growth factor levels are associated with vascular permeability39 and hence contrast enhancement, the presence of large regions of nonenhancing tumor in IDH1 mutants implies that this molecular genotype has low vascular endothelial growth factor levels, in contrast to a prior report.40 The frontal lobe predilection for IDH1 tumors is notable. There is some precedent for lobar specificity for molecular subtypes of glioma. For instance, in oligodendrogliomas, 1p19q-deleted tumors also appear to be more common in the frontal lobe,41 and some have suggested that insular oligodendrogliomas are rarely or never 1p19q-deleted,42 though this finding is controversial.43 GBMs have varying amounts of oligodendroglioma component, and this pathologic finding is associated with 1p19q deletions and better outcome.44,45 Thus it would be of interest to see if there is a relationship between an oligodendroglioma component with the 1p19q codeletion and the IDH1 mutation.
Our analysis is limited by its retrospective nature. In particular, our model of imaging features that can predict IDH1 mutational status will need to be confirmed in a prospective test set of patients. Also, it is possible that the relationship between imaging features and survival is dependent on treatment paradigms, which are constantly evolving. Steroid dose, which can affect edema and enhancement, was not available for many of our patients. Distinguishing edema from nonenhancing tumor can be challenging in some cases. Timing of contrast administration also could affect the classification of nonenhancing tumor because the proportion of tumor that enhances can increase as the time between contrast injection and scanning increases. However, most of the IDH1 mutants had large areas of nonenhancing tumor, potentially diminishing this limitation. Although the ability to identify nonenhancing tumor with high interobserver reliability has previously been demonstrated,46 the single-reader methodology used also is a limitation of this study. Approximately half of our patients received subtotal resection, and 12 had biopsy only. Therefore, it possible that sampling error could lead to underestimation of the IDH1 mutation incidence. However, it seems that in most patients, the IDH1 mutation tends to be present in all or most tumor cells, potentially mitigating the impact of this issue.24,25,47
Conclusions
We confirmed that MGMT promoter methylation and IDH1 mutation are associated with longer survival. Patients with methylated tumors with little or no peritumoral edema had particularly long survival. Standard MR imaging features may be able to accurately predict IDH1 mutational status but are poorly predictive of MGMT promoter methylation.
Footnotes
Disclosures: Albert Lai—RELATED: Grant: Roche,* Genentech,* Consulting Fee or Honorarium: Roche, Genentech, UNRELATED: Consultancy: Roche, Genentech, Grants/Grants Pending: Roche,* Genentech,* Merck,* Schering Plough.* Heidi Phillips—UNRELATED: Employment: Genentech, Stock/Stock Options: Genentech, Roche. Samir Kharbanda—UNRELATED: Employment: Genentech; Stock/Stock Options: Genentech. Timothy Cloughesy—UNRELATED: Consultancy: Genentech, Roche, Agios, Lilly, Novartis, Payment for Lectures (including service on Speakers Bureaus): Merck. Whitney Pope—UNRELATED: Consultancy: Genentech, Payment for Lectures (including service on Speakers Bureaus): Genentech, Payment for Development of Educational Presentations: Genentech. (*Money paid to the institution.)
References
- Received July 8, 2011.
- Accepted after revision October 11, 2011.
- © 2012 by American Journal of Neuroradiology