Abstract
BACKGROUND AND PURPOSE: Integration of imaging and genomic data is critical for a better understanding of gliomas, particularly considering the increasing focus on the use of imaging biomarkers for patient survival and treatment response. The purpose of this study was to correlate CBV and PS measured by using PCT with the genes regulating angiogenesis in GBM.
MATERIALS AND METHODS: Eighteen patients with WHO grade IV gliomas underwent pretreatment PCT and measurement of CBV and PS values from enhancing tumor. Tumor specimens were analyzed by TCGA by using Human Gene Expression Microarrays and were interrogated for correlation between CBV and PS estimates across the genome. We used the GO biologic process pathways for angiogenesis regulation to select genes of interest.
RESULTS: We observed expression levels for 92 angiogenesis-associated genes (332 probes), 19 of which had significant correlation with PS and 9 of which had significant correlation with CBV (P < .05). Proangiogenic genes such as TNFRSF1A (PS = 0.53, P = .024), HIF1A (PS = 0.62, P = .0065), KDR (CBV = 0.60, P = .0084; PS = 0.59, P = .0097), TIE1 (CBV = 0.54, P = .022; PS = 0.49, P = .039), and TIE2/TEK (CBV = 0.58, P = .012) showed a significant positive correlation; whereas antiangiogenic genes such as VASH2 (PS = −0.72, P = .00011) showed a significant inverse correlation.
CONCLUSIONS: Our findings are provocative, with some of the proangiogenic genes showing a positive correlation and some of the antiangiogenic genes showing an inverse correlation with tumor perfusion parameters, suggesting a molecular basis for these imaging biomarkers; however, this should be confirmed in a larger patient population.
ABBREVIATIONS:
- FGF2
- fibroblast growth factor 2
- GBM
- glioblastoma
- GO
- Gene Ontology
- MMP1
- matrix metallopeptidase 1 (interstitial collagenase)
- PCT
- perfusion CT
- PS
- permeability surface area product
- TCGA
- The Cancer Genome Atlas
- TNF
- tumor necrosis factor
- VASH1
- vasohibin 1
- VE
- vascular epithelium
- VEGF
- vascular endothelial growth factor
- WHO
- World Health Organization
GBMs account for 80% of malignant astrocytomas and are associated with poor prognosis despite aggressive multimodality combination therapies. Various imaging features have been used to characterize gliomas, beginning with contrast enhancement on conventional morphologic imaging and extending to the more advanced and complex metabolic and physiologic parameters by using various functional imaging techniques. However, within GBMs, there is marked inter- and intratumoral cytologic and histologic heterogeneity, which potentially leads to variable aggressiveness and prognosis. Imaging features—whether simple contrast enhancement and degree of necrosis or some of the quantitative measures such as perfusion parameters (tumor vascularity and angiogenesis), diffusion tensor estimates (tumor cellularity and invasiveness) and spectroscopic data, or PET metabolite uptake (tumor metabolism)—have been unable to robustly predict patient prognosis in this select subgroup.
Gene-expression profiling by using microarrays has demonstrated some of this intratumoral heterogeneity, revealing molecular signatures that reflect underlying pathogenic mechanisms and molecular features that may be associated with survival.1⇓⇓–4 The TCGA and Repository for Molecular Brain Neoplasia are extensive multidimensional datasets, which present a unique opportunity to integrate imaging and genomic data, which, in turn, provides unique opportunities for developing a more sophisticated understanding of gliomas.5 Considering the increasing focus on the use of quantitative imaging biomarkers for patient survival and treatment response, it is critical to understand the molecular basis of these imaging features. Recent literature has tried to correlate morphologic imaging features with gene expression in GBMs6⇓⇓–9; however, there has been little emphasis on correlating metabolic or physiologic imaging biomarkers with gene expression.
The purpose of this project was to retrospectively correlate hemodynamic (tumor blood volume) and physiologic (tumor vascular leakiness) perfusion parameters, which are surrogate imaging markers of tumor angiogenesis, with the expression of genes regulating angiogenesis in GBMs.
Materials and Methods
Our Health Insurance Portability and Accountability Act–compliant retrospective study was approved by the institutional review board (IRB #6381). This analysis included 18 patients with WHO grade IV gliomas (GBMs) who underwent pretreatment PCT between May 2005 and September 2009 and in whom tumor specimens were analyzed as part of TCGA project.10 Mean age was 57.7 years (range, 22–81 years) with 13 men and 5 women. All the patients underwent surgical resection (gross total resection, n = 4; subtotal resection, n = 14), and tumor specimens were collected as specified by TCGA.10 According to TCGA requirements, the pathology was confirmed as GBM with adequate frozen tissue ≥0.5 g with ≥70% tumor nuclei and <50% necrosis.
PCT Technique
Perfusion studies were performed by using 64-section multidetector row CT scanners (VCT; GE Healthcare, Milwaukee, Wisconsin) in all patients. A low-radiation-dose noncontrast CT head study was performed to localize the ROI before obtaining a perfusion scan. For the perfusion scan, 50 mL of nonionic contrast (ioversol, Optiray, 350 mg/mL; Mallinckrodt, St. Louis, Missouri) was injected at a rate of 4 mL/s through a 20-ga IV line by using an automatic power injector. At 5 seconds into the injection, a cine (continuous) scan was initiated with the following technique: 80 kV(peak), 120 mA, and 1 second per rotation for a duration of 50 seconds. After the initial 50-second cine scan, 8 more axial images were acquired for each section location, 1 image every 15 seconds for an additional 2 minutes, thus giving a total acquisition time of 199 seconds.11 Eight 5-mm-thick axial sections were acquired, resulting in a total coverage area of 4 cm.
PCT Map Analysis
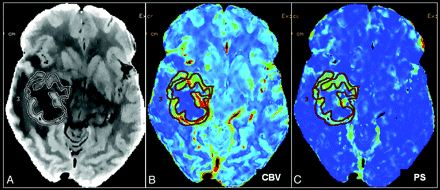
Perfusion maps of CBV and PS were generated at an Advantage Windows workstation by using PCT 3.0 software (GE Healthcare) and a 2-compartment model in all patients by a neuroradiologist with 9 years of experience. We used the superior sagittal sinus as the venous output function in all patients and the artery with the greatest peak and slope on time-attenuation curves as the arterial input function. An ROI was drawn within the confines of a large vessel, and the automatic function of the software selected the pixels with greatest peak and slope on the time-attenuation curve for analysis. ROIs were drawn manually on the PCT parametric maps, including the whole lesion within the coverage region. We placed ROIs to include the whole enhancing part of the tumor, taking care not to include necrotic/cystic parts or calcified portions of the lesion and also avoiding any major cortical vessels (Fig 1). Mean absolute values of CBV and PS in each case were used for the final analysis.
Perfusion CT maps. Base image (A), CBV (B), and PS maps (C) show placement of a freehand ROI, including the solid enhancing part of the tumor and excluding any cystic/necrotic parts.
TCGA Analysis
A custom 244 K Human Gene Expression Microarray (G4502A; Agilent Technologies, Santa Clara, California) was used to assess 18,407 genes in the human genome. The samples were processed by using the Universal Human Reference RNA pool (Stratgene, La Jolla, California). Data were preprocessed, and normalized within sample by locally weighted scatter plot smoothing as described elsewhere.10 These preprocessed level 2 data were obtained from the TCGA Data Portal.12 The data were further scale-normalized between arrays to reduce potential batch effects.13 Analysis was conducted on the probe level because probes within a gene may not behave homogeneously due to differences in hybridization. Clinical data were obtained from local patient records.
Statistical Analysis
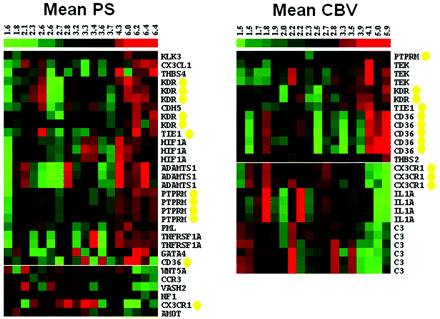
The Pearson correlation coefficient between the radiologic measure and the expression of each probe was estimated. Genes associated with the GO terms “positive regulation of angiogenesis” (GO:0045766) and “negative regulation of angiogenesis” (GO:0016525) were extracted for further analysis.14 Given the preliminary nature of the study and our small sample size, we did not adjust for multiple testing. The heat maps in Fig 2 are based on hierarchic clustering of the per-gene mean-centered gene expression by using average linkage (Gene Cluster 3.0)15 and visualization by using TreeView.15 The sample order was fixed according to increasing radiologic measure, and the column summary bar was colored to reflect this increase from low (green) to high (red).
Expression map showing genes with significant correlation ordered by hierarchic clustering with samples ordered by increasing PS (left) and CBV (right). Gene-expression levels are indicated relative to the probewise mean expression (black), with higher values in red and lower values in green. The summary bar at the top of the figure uses a similar color scheme to denote the level of the perfusion parameter. Genes with significant probes in both lists are highlighted with yellow circles.
Results
The gene expression data from TCGA were interrogated for correlation with CBV and PS estimates across the entire genome. There were 18,407 named genes represented by 55,328 probes. Of these named genes, 2910 (15.8%) were found to have a significant Pearson correlation coefficient at P < .05 with PS and the expression of at least 1 probe (4908 total probes). Additionally, 1137 genes (6.2%) had a significant correlation with CBV and the expression of at least 1 probe (1579 total probes). In common were 363 genes (9.9% of those significantly correlated).
Given our interest in CBV and PS estimates, we considered the GO biologic process pathways for angiogenesis regulation only. We observed expression levels for 92 genes (332 probes) in the regulation of the angiogenesis pathway (GO:0045766, GO:0016525), of which 19 genes (31 probes) had significant correlation with PS (Fig 2 and On-line Table 54,16⇓⇓⇓⇓⇓⇓⇓⇓⇓⇓⇓⇓⇓⇓⇓⇓⇓–34), whereas 9 genes (26 probes) had significant correlation with CBV (P < .05) (Fig 2 and On-line Table 621,22,24,27,31,35⇓⇓⇓–39). Five genes showed significant correlation with both CBV and PS (Fig 2).
To identify potentially spurious associations, we defined a set of criteria by which we deemed an association result to be consistent for a given gene. Genes with significant correlation coefficients (P < .05) of the same sign for least 2 probes were assumed to be “consistent.” When only 1 probe was significantly correlated, we looked for support from the remaining probes either with marginal significance resulting in at least 66% of the probes with P < .1 or with all probes exhibiting the same sign and a minimum absolute correlation coefficient of 0.3. Genes not meeting these criteria are noted as “not consistent” and should be interpreted cautiously given the present data (On-line Tables 5 and 6). One of 9 CBV-correlated genes and 9 of 19 PS-correlated genes failed to meet the requirements for consistency. See the supplemental material for correlation estimates of all 92 angiogenesis-related genes (332 probes).
Consistent positive correlation was found for 7 genes with PS and for 5 genes with CBV. The maximum absolute correlation coefficient with the associated P value is given for some of these genes: KDR VEGFR-2 (CBV = 0.60, P = .0084; PS = 0.59, P = .0097), HIF1A (CBV = 0.29, P = .29; PS = 0.66, P = .008), TNFRSF1A (CBV = −0.23, P = .3673; PS= 0.53, P = .0239), and TIE members TIE1 (CBV = 0.54, P = .0217; PS = 0.49, P = .0389), and TIE2/TEK (CBV = 0.58, P = .0119; PS = 0.46, P = .0550). Several genes showed marginal positive correlation (P < .1) to 1 measure which, given the sample size, is worth noting—ie, MMP9 (CBV = 0.42, P = .0883), ECM1 (CBV = 0.42, P = .0573), FGF2 (PS = 0.44, P = .0673), and Serpine1 [Serpin peptidase inhibitor clade E(nexin, plasminogen activator inhibitor type 1), member 1] (PS = 0.42, P = .0814).
Consistent negative correlation was found for 3 genes with PS and for 3 genes with CBV. The maximum absolute correlation coefficients with associated P values for some of these genes are VASH2 (CBV = −0.35, P = .1568; PS = −0.71, P = .0011), CX3CR1 (CBV = −0.66, P = .0028; PS = −0.49, P = .0375), WNT5A (CBV = −0.10, P = .6833; PS = −0.52, P = .0284), and C3 (CBV = −0.63, P = .0051; PS = −0.41, P = .0953).
Discussion
Malignant gliomas are known to be heterogeneous tumors in terms of their histologic features, angiogenesis, prognosis, and imaging features as well as their molecular and genetic composition. Recent developments in the microarray analysis of cancer genomes have led to an additional interest in the correlation of gene expression with imaging features in gliomas to better understand the physiologic basis for the imaging heterogeneity of these aggressive neoplasms. The limited number of publications on this topic has correlated the presence or absence of contrast enhancement with various gene-expression pathways affecting tumor cell mitosis, migration, angiogenesis, hypoxia, edema, and apoptosis.6⇓⇓–9 All except 1 focused on morphologic imaging features, and Barajas et al,9 though correlating histologic features with ADC and relative CBV estimates, did not do a direct correlation of physiologic measures with gene expression.
Tumor blood volume and permeability or leakiness estimates obtained by using dynamic contrast-enhanced imaging techniques (MR imaging or PCT) have been shown to correlate with tumor grade, prognosis, and treatment response.10,40⇓⇓⇓–44 These parameters have been shown to be imaging surrogate markers of tumor vascular attenuation and angiogenesis in gliomas,45 emphasizing their role as imaging biomarkers. In the present study, rather than focusing on morphologic imaging features, we chose to correlate the perfusion parameters (CBV and PS) obtained from the solid enhancing tumor directly with genes related to angiogenesis regulation.
Tumor angiogenesis is a critical and complex multistep process24,46⇓–48 for tumor progression, which is regulated by multiple pathways. Identifying key targets in pathways involved in the regulation and promotion of angiogenesis has resulted in the development of numerous therapeutic agents.49 The most prominent and characterized proangiogenic pathway is the VEGF signaling pathway. However, targeting 1 specific pathway has led to the development of resistance to some of these agents due to the continued evolving nature of these aggressive tumors, with tumor cells enabling other signaling pathways to continue their growth and invasiveness. Most of these pathways are regulated by a complex interaction of multiple genes, and some of these have been studied in detail with a much better understanding; however, there is no clear set of genes responsible for positive or negative regulation of angiogenesis. We used the genes listed in the GO biologic process pathways (GO:0045766, GO:0016525) for angiogenesis regulation and performed a literature search for individual genes that show significant correlation.
Correlation of Expression of Proangiogenic Genes with Perfusion Parameters
Among the proangiogenic genes that showed significant positive correlation with either 1 or both perfusion parameters are KDR (VEGFR-2), HIF1A, TNFRSF1A, and TIE1 and TEK/TIE2, which represent several pathways involved with angiogenesis. Of all the proangiogenic molecules, the most prominent and best-characterized is the VEGF signaling pathway, a key component in both the early and delayed phases of angiogenesis.49 VEGFR-2 (also known as KDR, Flk1), a member of the VEGF receptor family, is prominently expressed in the polarized extension of tip cell filopodia, enabling migration along the chemoattractant gradient.24 HIF1A is associated in both GBM stem and nonstem cells, with the induction of VEGF expression by transcriptionally regulating the VEGF promoter.23 Members of the TNF ligand superfamily and their receptors regulate various cellular responses including proliferation, migration, differentiation, and apoptosis.50 TNFRSF1A is 1 of the major receptors for the TNF-alpha. TEK/TIE2 signaling can aid in the recruitment of pericytes.38 Pericytes play an important role during the later stages of angiogenesis by stabilizing the maturing blood vessels, supporting endothelial cell survival, and also releasing proangiogenic molecules.51 The TIE family of receptor tyrosine kinases, TIE1 and TEK/TIE2, is activated by VEGF.27
Considering also those genes with marginal significance, we see the angiogenesis pathways expanded. MMP9 showed a borderline significant correlation with CBV but not with PS. MMP9 is known to be secreted by endothelial cells52 and is involved in the degradation of the basement membrane of endothelial cells, thus facilitating the movement of proliferating vascular sprouts into surrounding stroma and hence promoting angiogenesis.53 ECM1—also marginally correlated with CBV—is a proangiogenic protein produced by tumor cells, which stimulates the proliferation of endothelial cells and promotes blood vessel formation.54 FGF2, another key proangiogenic factor and a member of the heparin-binding protein family, was marginally correlated with PS but not CBV. FGF2 signaling can modify angiogenesis independent of the VEGF stimulus.55
What is surprising, a few genes such as CX3CR1 and WNT5A, which are presumed to be proangiogenic, showed a counterintuitive inverse correlation with perfusion parameters for which we do not have a clear explanation. CX3CR1 has been placed in the negative regulation of angiogenesis by the GO biologic process pathways list, though it is a receptor for CX3CL1, which is proangiogenic.21 However, CX3CR1 showed a consistently significant inverse correlation with both perfusion parameters, whereas CX3CL1 showed an inconsistent positive correlation with PS, which is likely spurious. WNT5A, a member of the WNT family of signaling proteins, is associated with the WNT/Ca(++) pathway, which has been shown to promote angiogenesis with induction of MMP1 and TIE2.18 However, other studies have found that WNT5 inhibits angiogenesis19 and may play a role in migration and infiltration in gliomas.20 Although seemingly counterintuitive, this inverse correlation between WNT5A and PS may be interesting if it can be validated.
Another interesting finding comes from the very high correlation of PTPRM with PS, consistent with a weaker correlation with CBV. Little is reported about this molecule with respect to angiogenesis, though it is included among the negative regulators of angiogenesis in GO. However, PTPRM has been shown to localize in the junctions between endothelial cells in lung tissue, leading to tighter junctions with increased expression.22 In this same study, PTPRM was found to interact with VE-cadherin (CDH5), which is known to play a role in tumor-associated angiogenesis and was also found to be positively correlated with PS.
Correlation of Expression of Antiangiogenic Genes with Perfusion Parameters
Among the genes related to antiangiogenic factors, VASH2 and C3 showed a significant inverse correlation with either or both perfusion parameters. VASH2 is an angiogenesis inhibitor,16 which showed an inverse correlation with PS estimates. However, another recent publication reported that VASH1 is expressed in endothelial cells in the termination zone to halt angiogenesis, whereas VASH2 is expressed in infiltrating mononuclear cells in the sprouting front to promote angiogenesis.17 In this study, we found no evidence of correlation with PS or CBV and VASH1. C3, though listed among the positive regulators of angiogenesis in GO, has been shown to participate in complement-mediated inhibition of neovascularization. This is compatible with the consistent strong negative correlation of C3 with CBV and the marginally negative correlation with PS.
AMOT and NF1, both negative regulators of angiogenesis, show negative correlations with PS, though the probe-level evidence does not achieve our definition for consistency. AMOT, a negative regulator of angiogenesis and permeability, is a receptor for the angiogenesis-inhibitor angiostatin, which regulates endothelial cell migration and tube formation. (Angiomotin regulates endothelial cell-cell junctions.30) NF1 has antiangiogenic properties as evidenced by increased expression of proangiogenic factors and decreased expression of antiangiogenic factors in neurofibromin-deficient Schwann cells. NF1 deletion has also been identified in the mesenchymal subtype of GBM,4 and it showed an inverse correlation with PS estimates, confirming an antiangiogenic effect.
Many of the other antiangiogenic genes, such as CD36, THBS2, THBS4, PML, KLK3 (PSA), and ADAMTS1, showed a positive correlation with perfusion parameters. Except for CD36 and ADAMTS1, these genes failed to meet our criteria for consistency. Thus, it is likely that most of these unexpected results are spurious. ADAMTS1 is known to be an angiogenesis inhibitor; however, VEGF significantly induces ADAMTS1 expression in endothelial cells, and this may represent a mechanism for feedback of angiogenesis inhibition.28 This mechanism may possibly account for the counterintuitive relationship in our study due to presumably increased VEGF activity as proved by increased expression of KDR. CD36 is also a receptor, accepting THBS1, which is a potent inhibitor of angiogenesis. THBS1 is not significantly correlated with PS or CBV in our study; this finding may suggest that overexpression of CD36 is also related to a negative feedback loop that is disrupted. These regulatory hypotheses need further study.
Limitations of the Study
As noted above, the small sample size of our study reduces the power to detect other potentially meaningful associations. The small sample size also prohibits subgroup analysis. Determining how these perfusion parameters and associated gene changes relate to currently proposed molecular subclassification of GBMs3,4 is a logical next step when a larger set of samples is obtained. Another limitation when performing this kind of analysis is a lack of clearly defined pathways for angiogenesis, even though we used the GO reference.14 In the present study, we correlated perfusion parameters obtained from the entire solid enhancing part of the tumor with microarray analysis done from the tumor, which was obtained during surgical debulking, which may not be representative of the solid enhancing part. However, TCGA tumor-specimen requirements do force the participating centers to submit mostly solid parts of tumor with little necrosis. Furthermore, at our center, neurosurgeons aim for safe maximal resection of the enhancing part in GBMs. We are currently proposing a study that will obtain image-guided biopsy specimens with a much better correlation of biopsy site–specific tumor perfusion parameters with microarray analysis done from this obtained biopsy specimen.
Conclusions
Although our study is limited by small sample size, the data show that CBV and PS estimates in GBMs can correlate positively with proangiogenic genes and inversely with antiangiogenic genes; this correlation can help establish a genomic/molecular basis for these commonly used imaging biomarkers and potentially adds to our knowledge of their immunohistologic bases.45 Our results indicate that further investigation with a larger study number is warranted and that correlation of imaging/perfusion parameters such as CBV and PS estimates obtained in vivo with genomic profiles could potentially lead to their use as surrogate imaging markers for gene expression–based classification of gliomas.
Acknowledgments
We thank Susan MacPhee-Gray for her help with manuscript editing and reformatting. We also thank the In Silico Glioma Phenotype Research Group5 for their invaluable help with facilitating genomic data retrieval.
Footnotes
Disclosures: Sandra Rempel—UNRELATED: Grants/Grants Pending: National Institutes of Health,* Payment for Lectures (including service on Speakers Bureaus): Department of Neurological Surgery, Center for Molecular Neurobiology, Ohio State University, April 2011. (*Money paid to the institution.)
Paper previously presented at: 49th Annual Meeting of American Society of Neuroradiology, June 4–9, 2011; Seattle, Washington.
References
- Received August 2, 2011.
- Accepted after revision October 11, 2011.
- © 2012 by American Journal of Neuroradiology