Abstract
BACKGROUND AND PURPOSE: Dysfunction of DA neurotransmission from the SN and VTA has been implicated in neuropsychiatric diseases, including Parkinson disease and schizophrenia. Unfortunately, these midbrain DA structures are difficult to define on clinical MR imaging. To more precisely evaluate the anatomic architecture of the DA midbrain, we scanned healthy participants with a 7T MR imaging system. Here we contrast the performance of high-resolution T2- and T2*-weighted GRASE and FFE MR imaging scans at 7T.
MATERIALS AND METHODS: Ten healthy participants were scanned by using GRASE and FFE sequences. CNRs were calculated among the SN, VTA, and RN, and their volumes were estimated by using a segmentation algorithm.
RESULTS: Both GRASE and FFE scans revealed visible contrast between midbrain DA regions. The GRASE scan showed higher CNRs compared with the FFE scan. The T2* contrast of the FFE scan further delineated substructures and microvasculature within the midbrain SN and RN. Segmentation and volume estimation of the midbrain SN, RN, and VTA showed individual differences in the size and volume of these structures across participants.
CONCLUSIONS: Both GRASE and FFE provide sufficient CNR to evaluate the anatomy of the midbrain DA system. The FFE in particular reveals vascular details and substructure information within the midbrain regions that could be useful for examining structural changes in midbrain pathologies.
Abbreviations
- AP
- antero-posterior
- BOLD
- blood oxygen level–dependent
- CNR
- contrast-to-noise ratio
- DA
- dopamine or dopaminergic
- EPI
- echo-planar imaging
- FA
- flip angle
- FFE
- fast-field echo
- FH
- foot-head
- fMRI
- functional MR imaging
- GRASE
- gradient and spin echo
- RL
- right-left
- RN
- red nucleus
- RNdv
- red nucleus densely vascularized section
- RNpv
- red nucleus poorly vascularized section
- SENSE
- sensitivity encoding
- SN
- substantia nigra
- SNc
- substantia nigra pars compacta
- SNR
- signal-to-noise ratio
- SNr
- substantia nigra pars reticulata
- SWI
- susceptibility-weighted imaging
- VTA
- ventral tegmental area
The DA neurotransmitter system arises from within the midbrain SNc and the VTA and projects to a broad range of cortical and subcortical brain regions.1–3 The DA neurons of the VTA and SNc have unique firing properties4 known to be associated with behaviors (in rodents and nonhuman primates) such as reward related learning,5 motivation,6,7 attention,8 novelty processing,9 and other goal-directed functions.10 While these in vivo studies in rodents and nonhuman primates provide a framework for understanding the role of the VTA and the SNc, the finer, distinct, and individual functions of these 2 nuclei in mediating behavior, especially in humans and in disease states, are still to be fully elucidated.
Accurately identifying the anatomy of midbrain DA regions in humans is essential for evaluating structural changes associated with neurodegenerative diseases. Loss of DA neurons in the SNc is known to play a causal role in the development of parkinsonian symptoms.11 Moreover, neuroimaging studies in drug addiction,12 schizophrenia,13 and psychosis14 have reported disease-related differences in BOLD activity in the VTA/SNc DA region, consistent with models of dysregulation of midbrain DA circuitry in these disorders.
Most MR imaging studies involving the brain stem to date have been conducted in scanners operating at 1.5T,15 3T,16,17 or 4T,18 with imaging protocols of limited resolution, sensitivity, and SNRs. However, at higher field strengths (≥7T), some of these limitations can be ameliorated because higher SNR allows the use of smaller voxel dimensions, and susceptibility contrast effects increase with increasing field strengths. MR imaging performed at 7T has previously been used to obtain higher resolution anatomic detail of brain structures19 and thus can be especially suitable for imaging regions such as the midbrain SN and VTA. To the extent that these areas can be delineated, it is possible to measure volumetric changes in these regions in disease states. Moreover, the magnetic susceptibility associated with neuromelanin, ferritin, and other iron-bearing compounds20 in mesencephalic structures such as the SN allows changes in deposits of these structures to be visualized and quantified. This finding is specifically relevant for clinical studies because levels of neuromelanin, iron, and ferritin all change during aging and Parkinson disease.21–23 Successfully imaging the midbrain structures may thus be useful for the detection and accurate diagnoses of neurodegenerative disorders pertaining to these regions.
T1-, T2-, and T2*-weighted pulse sequences have each been used to study midbrain anatomy, with T1-weighted sequences showing detectable contrast for lower midbrain structures.24 However, neither T1-weighted nor proton attenuation−weighted imaging has been able to delineate areas with different levels of vascularization within the RN or to detect subregions of the SN (pars compacta and reticulata). T2- and T2*-weighted imaging techniques at 3T and 4T have been able to obtain high-resolution anatomic images at 0.5 × 0.5 × 2 mm3 in reasonable imaging times with contrast revealing some vascular definition.18,25 However, MR imaging at these lower field strengths has typically been limited in its ability to accurately detail boundaries or detect the substructure and microvasculature of mesencephalic regions. Imaging at higher field strengths (such as 7T) with higher resolution could allow a more accurate examination of the anatomy of the midbrain SN and VTA.
The current study aims at evaluating the ability of high field (7T) MR imaging to scan the anatomic architecture of the midbrain DA regions by using T2- and T2*-weighted pulse sequences, including GRASE and 3D gradient-echo (also called FFE) sequences. GRASE26 is a T2- and T2*-weighted multishot imaging sequence comprising refocusing pulses coupled with trains of alternating polarity readout gradient lobes to form a series of interleaved spin and gradient echoes. This scan acquires data in 2 dimensions, with phase-encoding in 1 direction, allowing SENSE acceleration in 1 dimension. The FFE27 is a T2*-weighted multishot gradient-echo scan that acquires data in 3D. These sequences were chosen on the basis of considerations of imaging time, contrast sensitivity, and relative immunity to image distortions that can arise at high fields.
Using GRASE and FFE sequences, we obtained high-resolution images of the midbrain to detect visible susceptibility differences in the SN, VTA, and RN and to determine the ability of these sequences to observe substructure details. Quantitative assessments of CNR between different midbrain regions were computed. Additionally, a segmentation algorithm28 was used to segment the SN, VTA, and RN to create labeled volume maps and to estimate volumes of these midbrain regions across subjects.
Materials and Methods
Ten healthy volunteers, 20–40 years of age (7 men), were imaged on a 7T Achieva Scanner (Philips Healthcare, Cleveland, Ohio) by using both GRASE and FFE pulse sequences. All participants gave written informed consent for the study, which was approved by the institutional review board of Vanderbilt University Medical Center. Images were acquired in an oblique transverse orientation with 14 sections positioned to cover the dorsoventral extent of the midbrain from the inferior edge of the basal ganglia to the dorsal border of the pons. The transverse sections were tilted to make them parallel to a plane bisecting the mammillary body and the superior colliculi. See On-Line Fig 1 for section positioning information.
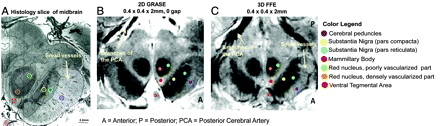
GRASE and FFE scans in a single subject next to a histology section of the midbrain at the level of the superior colliculus. A, Histology section of the midbrain (stained by using India ink to highlight vasculature). Reprinted with permission from Duvernoy.31 B, GRASE image of the midbrain. C, FFE image of the midbrain. The sections are oriented so that the anterior part of the midbrain appears at the bottom of the sections. Legend indicates the location of the midbrain structures marked with respect to a brain atlas.31
Imaging Protocol at 7T
After initial exploration of different sequence parameters, 2 different scanning sequences were used to image the anatomy of the midbrain. A flow-compensated 2D GRASE sequence was performed with a 0.4 × 0.4 mm in-plane resolution, a 2-mm section thickness, and a 0-mm section gap. Other scanning parameters included the following: FOV = 128 (AP) × 102 (RL) × 28 mm (FH) with foldover suppression, reconstruction matrix size = 320 × 256, TR = 5000 ms, TE = 22 ms, FA = 79°, SENSE or parallel imaging factor = 2 in the RL direction, turbo spin-echo factor = 4, EPI factor = 3, and acquisition time = 11 minutes 12 seconds. Within this time, 2 volumes were reconstructed, each with NEX = 3.
The FFE sequence was also flow-compensated, obtained at a resolution of 0.4 × 0.4 mm in-plane with 2-mm section thickness. Other scanning parameters included the following: FOV = 128 (AP) × 102 (RL) × 28 mm (FH) with foldover suppression, reconstruction matrix size = 320 × 256, TR = 25 ms, TE = 18 ms, FA = 15°, SENSE or parallel imaging factor = 2 in the RL direction, and acquisition time = 5 minutes 26 seconds. Again, 2 complete volumes, each with NEX = 3, were obtained.
Data Analysis
The data were processed by using Matlab (MathWorks, Natick, Massachusetts), and statistical tests were performed by using the Statistical Package for the Social Sciences, Version 18.0 (SPSS, Chicago, Illinois). Window-level-setting adjustments were made to view the best image contrast within the midbrain. To clearly visualize the different midbrain structures and improve image quality, we averaged the 2 volumes for each scan type together.
CNR Measurements
For GRASE and FFE scans, contrast measurements were calculated by taking the average of 2 volumes and measuring the signal intensity for a given structure by manually tracing out the volume of the structure in 2 dimensions. This calculation was performed across all the imaging sections. CNR was calculated as the signal-intensity difference between 2 structures divided by the noise. Noise was calculated by subtracting 1 image volume from the other and computing the pooled SD of the signal intensity in the 2 structures within the difference image. This process helped control for SENSE reconstruction artifacts. The calculation of the average and difference images assumes that the participants did not move their heads between the acquisitions of each volume.
Segmentation
A region-growing segmentation algorithm28 was used to trace out the boundaries of the midbrain structures. This algorithm uses the average image (produced from the 2 volumes) for every section, normalizes the signal intensity values to account for magnetic field inhomogeneities, and traces the contours of the tissue/structure by comparing the signal-intensity changes from 1 voxel to the next. All voxels above a certain intensity threshold are counted as part of 1 structure, and after 300 iterations of such intensity-threshold comparisons, the boundary of the structure is traced. This method worked very effectively for the SN and the RN, both of which had a sharp contrast between their boundaries and neighboring regions, presumably due to the signal-intensity loss associated with the magnetic susceptibility produced by iron products within their cells.
The SN was chosen from the inferior transverse section, just above the pons where the interpeduncular fossa opens up to the interpeduncular cistern. The most superior transverse section containing the SN includes a small portion of the posterior commissure, with the posterior boundary marked by the cerebral aqueduct and the anterior boundary defined by the mammillary bodies. The SN runs into the subthalamic nucleus in its dorsal boundary, so we used an atlas29 to visually guide the contours of the SN in the superior sections. We marked the SN in every section going in the superior direction until the transverse sections did not include the superior colliculi or the cerebral aqueduct. The subthalamic nucleus becomes prominent when viewed in the transverse plane that includes the third ventricle, RN, and posterior commissure with no superior colliculi.
The area of the VTA was manually traced and approximated in each section by localizing its boundaries with respect to neighboring regions as predicted by atlases.29,30 Similar to the SN, the VTA was chosen from inferior transverse sections, just above the pons, where the interpeduncular fossa opens up to the interpeduncular cistern. In these inferior sections, the boundaries of the VTA were defined with the lateral portion adjacent to the SN, and the medial portion adjacent to the interpeduncular fossa, abutting the ventral part of the superior cerebellar decussation and the interpeduncular nucleus. More superiorly, the VTA becomes a contiguous structure, with its lateral boundary still adjacent to the SN, and its medial boundary approaching the midline, apposing the interpeduncular fossa and extending in the posterior direction half way up the medial edge of the RN. This posterior boundary is a topographic landmark because the VTA lacks clear demarcation along the midline. The superiormost transverse section that contains the VTA includes a small portion of the posterior commissure, with the posterior boundary defined by the cerebral aqueduct and the anterior boundary defined by the mammillary bodies. The RN was chosen in all the sections that showed a visible contrast for this structure through the inferior-superior plane of axis. This region was traced as a control region to clearly distinguish and highlight the boundaries of the SN and VTA.
The segmentation algorithm was implemented across every section to produce a 3D-labeled volume map of the SN, VTA, and RN. The lateral sides of each structure were traced; this tracing segmented a total of 6 structures for every participant (left SN, left VTA, left RN, right SN, right VTA, and the right RN) and gave an estimate of the variation in the structure across the left and right sides. All the sections were visually inspected by overlaying the segmented maps onto the original anatomic image to ensure accuracy of overlap and successful segmentation.
Volume Measurement
Once the segmentation procedure was completed, the algorithm computed the number of voxels included within the segmented area in every section for every region of interest. The number of voxels was then multiplied by the size of each voxel (0.4 × 0.4 × 2 mm) to compute the volume of each structure in cubic millimeters.
Volume Normalization
To control for intersubject variability in midbrain size, we normalized each participant's volume of interest (SN, VTA, RN) to the entire midbrain volume. The midbrain volume was estimated by segmenting the entire midbrain in the GRASE scan for every section that included the segmented SN, VTA, or RN. We used this midbrain normalization rather than whole-brain normalization because it allows specific consideration of the extent to which the 3 structures of interest make up the midbrain region.
Results
Visible Contrast in Midbrain Regions
Both GRASE and FFE scans revealed visible contrast in the midbrain regions, with anatomic detail close to that observed in histology (Fig 1). The densely and poorly vascularized regions of the RN were distinctly visible in both the GRASE and FFE scans (Figs 1B, -C and 2). Additionally, minute vascular contrast was evident in the FFE scan for small blood vessels within the SN (Fig 1C). The FFE scan also displayed distinct contrast between subregions of the SN (pars compacta versus reticulata) showing sensitivity to iron-related magnetic susceptibilities (Fig 2).
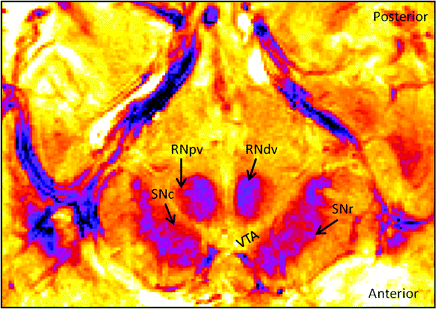
FFE image in a single subject in 1 section of the midbrain at the level of the superior colliculus. The image was passed through a lookup table filter to produce a color-contrast scale. Different parts of the RN and the SN are revealed.
CNR Measurements
Across the 10 subjects, GRASE and FFE scans showed detectable contrast between midbrain regions (Table 1).
Mean CNR values for GRASE and FFE scans in different midbrain regions across 10 participants
When we compared across subjects, the CNRs from the 2 scan types were not strongly correlated. The Pearson correlation coefficient of CNR values between the 2 different scan types was r = 0.47 for SN/VTA, r = −0.46 for the SN/RN, and r = 0.21 for the RN/VTA (all P > .10). CNR differences between the 2 scans varied depending on the specific ratio in question. No differences were observed across the GRASE and FFE scans between the SN and VTA, and the SN and the RN (both, Wilcoxon signed rank test, P > .10). However the CNR difference in the RN/VTA was significant across the 2 scans (Wilcoxon signed rank test, z = −2.70, P = .007). Additionally, a section-by-section comparison of the CNR values showed inter-section variability across subjects (On-line Fig 2).
Segmentation and Volume Measurements
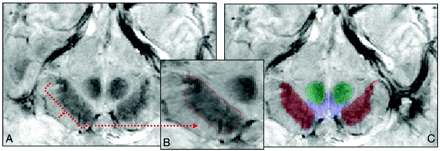
The segmentation algorithm traced the midbrain regions in both the GRASE and FFE scans (Fig 3 and On-line Fig 3). The area identified by the algorithm corresponded to all visually observable boundaries.
Segmentation process by using a region-growing segmentation algorithm28 in the FFE scan. A, Sample section in the midbrain at the level of the mammillary bodies where the segmentation algorithm was implemented. B, Segmented region of the SN traced in red within the midbrain. C, Segmented structures (SN is red; VTA, blue; and RN, green) overlaid on the anatomic FFE image.
From the segmented images, the volumes of different midbrain regions were computed (Table 2 and Fig 4).
Mean absolute and normalized volume estimates from GRASE and FFE scans in the SN, VTA, and RN
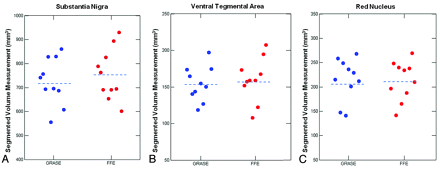
Individual-subject absolute-volume measurement for the SN (A), VTA (B), and the RN (C). Volume measurements for 10 subjects are shown here. Blue dots represent the GRASE scan; and red dots, the FFE scan. The dashed line represents mean volume.
The GRASE and FFE scans showed high correlation between the volumes of the midbrain regions (Pearson correlation coefficient for GRASE and FFE scans of the SN, r = 0.93; VTA, r = 0.87; and RN, r = 0.96 (all P < .001). Additionally, the GRASE and FFE volume results were not significantly different from each other (Wilcoxon signed rank test for the VTA, z = −0.97, P = .33; for the RN, z = −1.07, P = .29), though there was a modest trend for higher volumes by using the FFE over the GRASE sequence for the SN (z = −1.68, P = .09).
The volumes of the RN, SN, and VTA were normalized with respect to the midbrain volume to control for variability in the dimensions of structures in the midbrain across participants (Table 2 and On-line Fig 4). The GRASE and FFE scans again showed high correlation between the volumes of the midbrain regions (SN, r = 0.92; VTA, r = 0.81; and RN, r = 0.89; all, P ≤ .005). Additionally, the GRASE and FFE volume results were not significantly different from each other (all, Wilcoxon signed rank test, P > .10).
Volume results described above were averaged across the 2 sides to control for laterality differences. When laterality was taken into consideration, the SN and RN did not show a significant difference between the left and right sides (all, Wilcoxon signed rank test, P > .10). In contrast, the VTA showed a significant difference between the left and right sides in both the GRASE and FFE scans (GRASE, Wilcoxon signed rank test, z = −2.70, P = .007; FFE, Wilcoxon signed rank test, z = −1.99, P = .047).
Discussion
Anatomic Delineation Achieved at 7T
Our results indicate that both GRASE and FFE scans provide exquisite anatomic detail in the midbrain, revealing close concordance to structures highlighted in histology (Fig 1). The mixed T2- and T2*-weighting of the GRASE scan distinctly delineated the boundaries of the midbrain area, including iron-rich areas within the SN and RN. The T2*-weighting of the FFE scan was also comparable with the GRASE scan for detecting visible contrast among structures within the midbrain.
Vascular contrast was observed in both scans, especially within the RN, where the medial and lateral areas have different MR imaging signal intensities, corresponding to the densely and poor vascularized portions respectively and matching similar areas in the histologic section stained for vasculature (Fig 1). The vessel-related contrasts observed within the RN also corroborate previous work shown by using SWI in the midbrain at 4T.18 The T2*-weighting of the FFE scan could additionally detect subtle microvascular details, such as small blood vessels emanating from the SN complex, traveling toward the cerebral peduncles (Fig 1). These comprise multiple arteries and veins belonging to the internal anterolateral group of mesencephalic vessels, crossing through the cerebral peduncles at right angles to the peduncles.31 This degree of vascular definition achieved through MR imaging is especially beneficial in clinical settings during the assessment of vascular infarcts or ischemia in the midbrain or other brain areas connected to the midbrain.
The T2-weighting in both the GRASE and FFE scans, and in particular the latter, enabled the detection of signal-intensity variations within different parts of the SN, namely the SN pars compacta, SNc and SN pars reticulata (Fig 2). The SNr is known to have higher iron content compared with the SNc and has been shown to be correlated with decreased signal intensity (associated with decreased T2 relaxation times).32 The SNc contains more neuromelanin than the SNr,33,34 and iron can be stored in neuromelanin by the iron-storage protein ferritin.35,36 Iron alters the magnetic field uniformity in tissues and causes MR signal intensity within tissues to decrease.32,37,38 Hence the MR imaging−related signal intensity effects observed in the SN could be attributed to the paramagnetic neuromelanin or intracellular iron stores. In our study, we observed a characteristic signal-intensity difference in the SN complex, with a relatively hyperintense band framing the SNc and a relatively hypointense band framing the SNr (Fig 1C), showing delineation of these areas with imaging previously attempted using T2-weighted sequences at 1.5T.15
The color-coded image of the midbrain in the FFE scan (Fig 2) shows visible signal-intensity effects in the SN. This visual demarcation in signal intensity may represent the boundary of the SNc and SNr and was observed across sections. However, the reliability of the divide and its relation to the actual boundaries of the SNc remains uncertain. A clear delineation of the boundary of these 2 structures in humans by using MR imaging is a controversial matter. Manova et al18 point to a possible divide in the SN by using SWI without articulating the specific susceptibility difference in the 2 structures or providing details on the criteria for defining this divide. T1-weighted neuromelanin-sensitive scans17,24 have also been used to localize the SNc, without describing the boundary between the SNc and the SNr or discussing how the iron in the SNr may contribute to T1 shortening or lengthening effects.38
Other studies showed decreased signal intensity in their T2-weighted images, making reference only to the reticulata in their identification of SN32; or on the basis of signal intensity, they attributed the medial hypointense region to the SNr and the lateral higher signal intensity area to the SNc.39 Additional studies used an approach that appears to capture the intermediate area between the nigra and RN, rather than specifically the nigra.15,40,41 This finding implies that establishing clearly defined reliable boundaries between the reticulata and compacta has yet to be demonstrated in the literature. Comparison of postmortem samples and establishment of inter-rater reliability of the SNr and SNc volumes would be useful for verifying the boundaries of these substructures in future research.
Clinical Implications of Understanding the Anatomy of the Midbrain
Characterizing the anatomic architecture of the SN has important clinical implications, especially in the diagnosis of Parkinson disease, in which there is a known loss of DA neurons in the SNc11 and an increase in iron deposition within the SN.42 The source of MR imaging signal-intensity changes in Parkinson disease is not fully understood. Studies have shown a reduction in the width of the SNc, arising from an increase in iron content within the SNr or loss of DA neurons in the SNc.15 In MR imaging, these structural changes have been associated with a “smudging” of the SNr hypointensity along with decreased distance between the RN and the SN.43 Other studies have shown T2-weighted hyperintense signal intensity within the SNr or a loss of the hypointense signal intensity within this area.40,44 The region-based segmentation algorithm used in the present study provides a means of quantifying the dimensions and distance between various midbrain brain structures, especially at high-voxel resolutions.
This information can be further used to trace tissue-related structural changes occurring longitudinally. T2 relaxation time provides another marker for measuring disease-related structural changes within the SN45 with R2′ measurements in the SN at 3T shown to correlate with simple motor scores in Parkinson disease.46 Susceptibility contrast effects increase with increasing field strengths, so MR images like FFE and GRASE used in the present study at 7T should be particularly sensitive to subtle T2- and T2*-weighted changes seen in the anatomy of the SN in neurodegenerative diseases.
Quantitative Measurements within the Midbrain by using CNR and Volume Segmentation
The CNR measurements taken between different regions of the midbrain revealed that the CNR of the GRASE scans was slightly higher than that of the FFE scan (Table 1), though the scanning time was also longer. The CNR values achieved in the present study are higher than those reported by previous imaging studies of the mesencephalon at 4T,18 showing greater sensitivity to detecting variations in tissues at higher field strengths. Variability in the mean CNR values across subjects for various sections was also observed, suggesting region-specific differences in signal intensity and noise measurements.
Volume measurements of the SN, VTA, and RN showed individual differences, indicating variability based on the size of the structure, even after correcting for the overall size of the participant's midbrain. The volumes of the midbrain regions in this study were not normalized with respect to the total intracranial volume because we did not collect whole-brain data. This method of normalization could certainly be implemented in future studies. While we did not observe laterality differences for the SN and RN, volume differences were observed across the left and right VTA in both the GRASE and FFE scans. Unlike the SN and the RN, the VTA does not have any MR imaging−specific contrast that would aid in its detection. Thus, the boundaries of the VTA were manually defined by using anatomic reference points based on existing atlases.29,30 The positions of some of these reference points (such as the interpeduncular fossa) were estimated on the basis of the detectability of boundaries arising in the specific scan sequence. While the GRASE sequence clearly delineates the overall boundaries of the midbrain, these boundaries are less apparent in the FFE scan, contributing to the variation of volumes when defining the VTA region.
Within neuroimaging research, knowledge of midbrain boundaries can be beneficial for localizing BOLD responses in fMRI experiments of the midbrain. Due to imaging resolution limitations, most prior fMRI studies have not shown distinct and reliable BOLD activity in the SN and VTA for tasks thought to invoke DA release.47,48 Defining the precise boundaries of different midbrain regions can also be useful for localizing seed regions in functional connectivity maps within the brain stem.
Higher field strength imaging has various technical challenges, especially when imaging the midbrain. Some of these challenges include signal-intensity artifacts and distortions caused by large-scale magnetic susceptibility variations in the brain and the behavior of radio-frequency coils at high frequency. While the scan sequences described in the current work can handle some of these technical challenges, the development of more uniform B1 magnetic fields, increasing the sensitivity of parallel imaging by using a higher number of coil elements, and optimizing scan sequences for imaging-specific areas within the brain stem may provide a more tailored approach to imaging the midbrain in the future. SWI appears particularly promising at 4T; high-pass filtered phase images show MR imaging susceptibility contrast that may be capable of distinguishing different parts of the SN.18 In preliminary studies with SWI at 7T, we have observed some evidence of enhanced distinction between the SNc and SNr but have also observed phase-related aliasing at the boundary of the midbrain and the interpeduncular fossa (tissue-CSF interface) and lower CNR values for the SN as a whole in comparison with the magnitude images of T2*-weighted FFE and GRASE scans. Future studies at 7T in which optimized SWI data are combined with GRASE or FFE scans may provide further improvements in accurately delineating midbrain substructures.
Conclusions
In this study, we successfully imaged the midbrain SN and VTA in healthy humans to delineate anatomic detail that was otherwise not achievable at lower field strength MR imaging. The signal-intensity sensitivity was calculated by using CNRs, and the volume of the midbrain was estimated by segmenting individual midbrain structures. The results from this study not only further our understanding of the fine-grained anatomic architecture of the midbrain but also show potential clinical relevance in neuropsychiatric diseases in which there are known structural changes pertinent to this area of the brain.
Footnotes
-
This work was supported by the Biomedical Resources Partnership–Integrated Imaging of Brain Function at 7T (NIH grant R01EB000461).
-
Indicates article with supplemental on-line figures.
Indicates open access to non-subscribers at www.ajnr.org
References
- Received July 7, 2010.
- Accepted after revision September 7, 2010.
- Copyright © American Society of Neuroradiology