Abstract
SUMMARY: In this second article, we review the various strategies and applications that make use of reporter genes for molecular imaging of the brain in living subjects. These approaches are emerging as valuable tools for monitoring gene expression in diverse applications in laboratory animals, including the study of gene-targeted and trafficking cells, gene therapies, transgenic animals, and more complex molecular interactions within the central nervous system. Further development of more sensitive and selective reporters, combined with improvements in detection technology, will consolidate the position of in vivo reporter gene imaging as a versatile technique for greater understanding of intracellular biologic processes and underlying molecular neuropathology and will potentially establish a future role in the clinical management of patients with neurologic diseases.
Molecular imaging seeks to shed new light on both structure and function by creating images that directly or indirectly reflect specific cellular and molecular events (eg, gene expression) that can reveal pathways and mechanisms responsible for disease within the context of physiologically authentic and intact living subject environments.1 We and others have previously reviewed the factors contributing to the emergence of molecular imaging, the particular advantages of these approaches, and the general goals potentially achievable in biomedical research and clinical practice by adopting molecular imaging strategies.1–5 One of the subdisciplines in molecular imaging that is least familiar to clinical imaging specialists, and arguably one that holds future promise in neuroimaging, is reporter gene expression imaging.6,7 In the first article of this series, we reviewed the basic principles and recent technologic developments in reporter gene expression imaging in living subjects.8 In this second article, we review examples from the myriad experimental applications currently possible in molecular neuroimaging.
Experimental Applications in Molecular Neuroimaging Using Reporter Genes
Four broad categories of experimental applications for reporter gene imaging in the brain are as follows: gene marking of cells and viruses with reporter genes, imaging of gene therapies, imaging of transgenic animals carrying reporter genes, and imaging of more complex intracellular molecular events such as protein trafficking. Some details of recent examples of these applications are displayed in the Tables, with 1 or 2 representative examples of each application discussed in greater detail below. Of note regarding terminology, “xenografts” result from transplantation of cells or tissues from 1 species to another (eg, human cells into mice), and “orthotopic transplants” refer to grafting cells into the same body location/organ as that from which the cells are derived (eg, glioma cells delivered into brain).
Imaging Gene-Marked Cells
Gene marking may be used to track the behavior of almost any tissue.9 It is necessary to transfect cells stably with the imaging marker gene if they and their progeny are to be followed for their entire lifespan within the living subject. However, this assumes that minimal or no promoter attenuation or shutoff takes place. The latter can contribute substantially to a decline in transgene expression despite the constitutive nature of the promoter. In practice, transient transfection of cells suffices if these marked cells are to be imaged in a living subject for no more than approximately 7–10 days, depending on the cells in question and other parameters as well.1 In principle, gene-marker studies may be used to follow the behavior of almost any cell type in living subjects. In clinical practice, this has been mostly used with hematopoietic cells.9 However, in molecular imaging research, a variety of cells can be engineered to incorporate reporter genes. Usually, gene marking of cells that are static in 1 location (eg, subcutaneous tumor xenografts or orthotopic brain tumor implants) is used for first assessment and continued validation of reporter genes and their probes, for refining the technical aspects of molecular imaging signal-intensity detection from the brain, or for studying the behavior of the cells themselves within living subjects. This can be accomplished in 2 ways: ex vivo transfection of the cells in question with a vector containing an imaging cassette, followed by placement of these cells in a living subject or direct in vivo placement, usually via injection of the vectors carrying the reporter gene, as part of the recombinant genome of viruses, into the cells of interest within the body.
There are numerous examples of bioluminescence imaging of cells (especially cancer cells) that are mostly destined to remain static in the brain after ex vivo gene marking with imaging reporters and subsequent placement in living rodents (Table 1).10–23 A noteworthy advantage in these cancer models is that they create the opportunity for temporal evaluation of cancer biology in a noninvasive manner. Dynamic studies of xenograft growth and regression, either spontaneously or after therapy, can be performed. The enzymatic emission of light by firefly luciferase (Fluc) is adenosine triphosphate–dependent; therefore, only living metabolically active cells contribute to the signal intensity. A decrease in signal intensity occurs as cells die.
Recent examples of applications in molecular neuroimaging using gene marking (of static cells)
One of the earliest applications of reporter gene imaging of orthotopic mouse brain xenografts of rat 9L gliosarcoma cells gene-marked with Fluc was conducted by Rehemtulla et al.10 Intracerebral tumor burden was monitored over time by quantification of light emission and tumor volume by using bioluminescence imaging and MR imaging, respectively. There was excellent correlation (r = 0.91) between detected photons and tumor volume. A quantitative comparison of tumor cell kill, determined from serial MR imaging volume measurements and bioluminescence imaging photon counts following 1,3-bis(2-chloroethyl)-1-nitrosourea treatment, revealed that both imaging techniques yielded statistically similar cell kill values (P = .951). These results provide direct validation of bioluminescence imaging as a powerful and quantitative tool for the assessment of antineoplastic therapies in living animals.
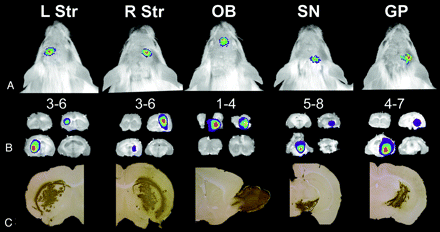
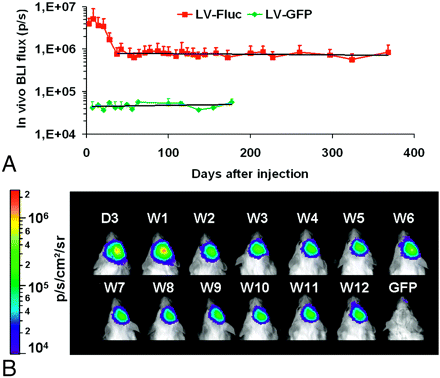
In a more recent study, Deroose et al11 reported the use of bioluminescence imaging to characterize lentiviral vector–mediated gene transfer into mouse brain. Various features of the imaging signals were characterized including localization (Fig 1), kinetics, resolution, and reproducibility. Although light signal intensity gradually decreased to 20% of initial values obtained in the first month, the signal intensity remained constant thereafter for more than 1 year after heterotopic brain xenografting of stably transduced 293T cells (Fig 2), allowing the potential for long-term evaluation of novel therapies for experimental brain disorders.
The localization of the bioluminescence imaging signal intensity reflects the anatomic site of injection. A, Bioluminescence imaging scans 14 days after injection of 293T cells transduced with a lentiviral construct encoding enhanced green fluorescent protein (EGFP) and Fluc separated by the internal ribosome entry site (I) of the encephalomyocarditis virus (LV-EGFP-I-Fluc) showing a focus that is located above the injection site: caudal to the eyes and on the left side of the head for the left striatum (L Str), caudal to the eyes and on the right side for the right striatum (R Str), between the eyes for the olfactory bulb (OB), near the caudal edge of the skull for the substantia nigra (SN), and intermediate between R Str and SN for the globus pallidus (GP). These sites correspond to the expected locations on the basis of the injection coordinates. B, Ex vivo bioluminescence images of 1-mm-thick coronal sections show the localization of the signal intensity at the site of injection. The sections are numbered in the anteroposterior direction from the bulbus olfactorius1 to the cerebellum.8 C, Immunohistochemistry for EGFP confirms the site of injection. (Reprinted by permission from Macmillan Publishers: Deroose et al. Mol. Therapy 2006;14:423–31, copyright 2006).
Time course of bioluminescence imaging (BLI) signal intensity after lentiviral (LV) transduction of mouse brain. A, Long-term evolution of the BLI signal intensity in a group of mice (n = 10) injected with 17-ng p24 of LV-Fluc and in a group injected with 8.4-ng p24 control vector (LV-enhanced green fluorescent protein [EGFP], n = 4). After a peak at days 8 to 14, the signal intensity declines during the first month to 16% of the maximum value at day 37 and then remains constant at 17.5 ± 2.3% of the maximum value from days 42 to 365. A linear regression line is drawn from days 37 to 365 (R2 = 0.027) for LV-Fluc and for all time points for LV-EGFP (R2 = 0.041). B, BLI of a representative animal shows an initial rise in signal intensity at week (W) 1 followed by a decrease and thereafter a stabilization of the signal intensity. The control animal shown represents the highest signal intensity seen in a control animal. GFP indicates green fluorescent protein; D, day; p, photons. (Reprinted by permission from Macmillan Publishers: Deroose et al. Mol. Therapy 2006;14:423–31, copyright 2006)
In vivo imaging of cell trafficking is currently performed in clinical practice (eg, by using indium-111 oxine for single-photon emission tomography [SPECT] imaging of infection and inflammation24) and is the objective of many immunologic and oncologic studies. Gene marking has the advantage over simple cell labeling for long-term tracking of cells because the imaging gene is passed on to the cell progeny and the imaging signal intensity is not lost through dilution by egress of the label from the cell.25 When a gene-marked cell dies or is phagocytosed by immune cells, the imaging signal intensity is also lost, unlike the situation with simple cell labeling in which the imaging signal intensity is not dependent on cell viability and may originate from extracellular space or from within immune scavenger cells.25
Table 2 outlines examples of recent reports that feature reporter gene imaging of trafficking virus particles,26–28 parasites,29 cancer cells,30 and stem cells to the brain.31–35 Viral marking is used mostly to study the pathogenesis of viral encephalitis, whereas marking of cancer cells may be used to investigate brain metastases from systemic primary sources (eg, breast cancer). An application gaining rapid acceptance in the laboratory is that of reporter gene imaging of neural stem cells to visualize, quantify, and time their trafficking to gliomas,31,32 ischemic brain,33,34 and injured spinal cord.35 Kim et al33 noninvasively imaged the migratory behavior of Fluc-marked neural progenitor cells to middle cerebral artery infarcts in mice and found that intraventricular delivery of stem cells results in earlier and more efficient infarct seeding. More recently, the same group showed that marked stem cells survived better in T-cell–deficient nude mice than in immunocompetent animals, indicating that immune status and host immunity can have an influence on stem cell graft survival in the cell therapy of experimental stroke.34
Recent examples of applications in molecular neuroimaging using gene marking (of trafficking cells)
Imaging of Gene Therapies
Although various methods of gene therapy have met with limited success, it is probable that eventually many diseases will be successfully treated with the delivery of 1 or more transgenes to target tissue. A concern in applying gene therapy is achievement of controlled and effective delivery of genes to target cells and avoidance of ectopic expression. Molecular imaging of reporters on particular therapeutic genes could be critical in optimizing gene therapy.36 The aim of these approaches is to image quantitatively reporter gene expression and from this to infer levels, location, and duration of therapeutic gene expression. There are several molecular strategies to achieve linkage of expression of the therapeutic transgene and the imaging reporter gene.37 These various techniques can be adopted with bioluminescence imaging reporter genes.
To date, several gene therapy studies have incorporated reporter gene imaging either to image the trafficking of the therapeutic transgene delivery vehicle alone,38 to image the target tissue in the brain alone (eg, to image the effect of therapy on an intracranial glioma by gene marking the glioma cells themselves),39,40 or when expression of the imaging gene is linked with that of the therapeutic gene to quantify transgene expression in the brain (Table 3).41 As an example of the latter, Rehemtulla et al41 developed an adenoviral vector containing both the therapeutic transgene yeast cytosine deaminase (yCD) along with Fluc. Following intratumoral injection of the vector into orthotopic 9L gliomas in rats, anatomic and diffusion-weighted MR images were obtained with time to provide for quantitative assessment of overall therapeutic efficacy and spatial heterogeneity of cell kill, respectively. In addition, bioluminescence images assessed the duration and magnitude of gene expression. MR images revealed significant reduction in tumor growth rates associated with yCD/5-fluorocytosine (5FC) gene therapy. Significant increases in mean tumor diffusion values were also observed during treatment with 5FC. Moreover, spatial heterogeneity in tumor diffusion changes were also observed, revealing that diffusion MR imaging could detect regional therapeutic effects due to the nonuniform delivery and/or expression of the therapeutic yCD transgene within the tumor mass. In addition, bioluminescence imaging in the living mice detected Fluc expression, which was found to decrease with time during administration of the prodrug, providing a noninvasive surrogate marker for monitoring gene expression. These results demonstrated the efficacy of the yCD/5FC strategy for the treatment of brain tumors and revealed the feasibility of using multitechnique molecular and functional imaging for assessment of gene expression and therapeutic efficacy.
Recent examples of applications of molecular neuroimaging in gene therapy
Imaging of Transgenic Models of Spontaneous Disease
The mouse is close to an ideal system to model human diseases because its genome can be easily manipulated and its anatomy and physiology are similar to that of humans. Combinatorial genetic engineering strategies to generate disease-prone genetic strains are now possible to produce new alleles by transgenic technology, in which extra deoxyribonucleic acid (DNA) that encodes the gene of interest is inserted heritably into the mouse genome or by knockout/knockin technology, in which specific portions of the mouse genome are targeted for selective alteration. For example, cyclization recombination (Cre) recombinase is an enzyme used to modify genes and chromosomes. A target region to be deleted in a gene locus can be marked for deletion by signal intensity sequences of locus of X-over P1 (loxP) that are identified by Cre. The expression of Cre leads to precise removal of the stretch of DNA between the recombinase signal-intensity sequences. With tetracycline-regulated and Cre recombinase–inducible alleles, the timing, duration, and tissue compartment of gene expression or inactivation can be further controlled. These technologies can be combined to yield faithful genetically modified mouse models of specific diseases that overexpress or lack genes of interest in all cells or only in a specific tissue compartment and/or developmental stage of interest.42
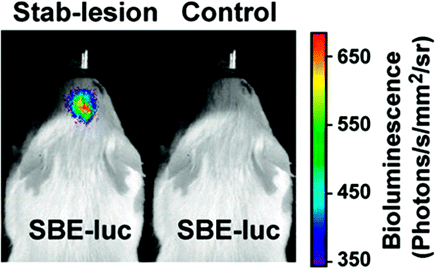
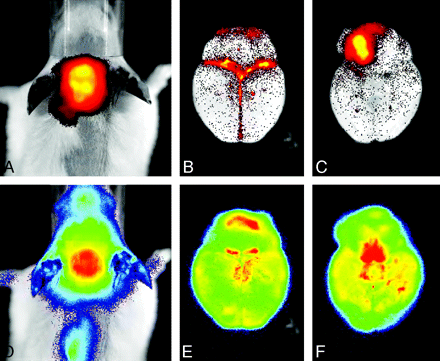
The strong merits of noninvasive imaging in the assessment of transgenic animals can be readily appreciated from the previous discussion of the overall advantages of molecular imaging in living subjects. More specifically, imaging techniques offer the possibility of adding in vivo phenotyping and monitoring of live animals for diagnostic purposes of disease identification, observing disease progression, and longitudinal effects of drug action, each mouse being its own control.43 To date, several research groups have used bioluminescence neuroimaging in their assessment of transgenic mice (Table 4).44–54 For example, Lin et al46 have imaged transforming growth factor (TGF)-β signaling in living mice in response to brain injury (Fig 3), and Kadurugamuwa et al51 have developed a method to simultaneously image pneumococcal meningitis and the accompanying neuronal injury (Fig 4). Other examples of transgenic models of spontaneous cancer, in which tumor formation is dependent on defined genetic alterations, provide a powerful test system for evaluating the therapeutic efficacy of pathway-specific antineoplastics. Vooijs et al54 have generated a conditional mouse model for retinoblastoma-dependent sporadic cancer that permits noninvasive monitoring of pituitary tumor development in living mice by bioluminescence imaging of Fluc expression. Bioluminescence imaging of pituitary cancer development with coexpression of the Fluc gene enabled longitudinal monitoring of tumor onset, progression, and response to therapy and may be used effectively for testing cancer prevention and treatment strategies on the basis of therapeutics that specifically target the retinoblastoma pathway. More recently, Momota and Holland55 have developed an imaging approach to measure cell proliferation in transgenic mice harboring gliomas with the E2F1 promoter (the E2F family are transcription factors) driving expression of Fluc. It is known that inactivation of the retinoblastoma protein (RB) pathway is one of the commonest alterations in gliomas as a result of mutations in either the RB gene or upstream regulators of phosphor-RB. The E2F1 promoter is strictly regulated by RB in cell-cycle progression and, in tumor cells, appears to mediate tumor-selective transgene expression. Linkage of the E2F1 promoter to Fluc allows the imaging of genetically induced loss of RB control as a model of human gliomas.
Brain injury results in activation of TGF-β responsive genes and the Smad-binding element (SBE)-firefly luciferase (luc) reporter. Two SBE-luc mice with similar basal levels of bioluminescence were lesioned with a needle stab to the right hemisphere or were left untreated (control), and bioluminescence was recorded 1 hour later. To highlight the increase in signal intensity in the lesioned mouse, the color scale was adjusted to leave the basal Fluc expression in the control mouse uncolored (<200 photons[p]/s/mm2/sr). (Copyright 2005, The American Association of Immunologists)
Simultaneous in vivo biphotonic monitoring of pneumococcal meningitis and the accompanying neuronal injury in live transgenic mice. Streptococcus pneumoniae engineered for bioluminescence (lux) was used for direct visualization of disease progression. Host response was monitored in transgenic mice containing an inducible firefly luciferase (Fluc) reporter gene under transcriptional control of the mouse glial fibrillary acidic protein (GFAP) promoter. On the basis of the different spectra of light emission and substrate requirements for Fluc and luc, it is possible to monitor separately the 2 reporters by using a highly sensitive in vivo imaging system. In vivo (A and D) and ex vivo (B, C, E, and F) images of brains from transgenic mice with meningitis were obtained at 19 hours postinfection. A, B, and C show lux imaging and D, E, and F show Fluc imaging. Dorsal and ventral views of an ex vivo brain show the bacterial and GFAP signals individually. Much of the bacterial signal intensity comes from discrete patches, whereas GFAP is induced in the entire brain and there are different intensities of the bioluminescence signal intensity in certain regions of the brain. Note the strong bacterial signal intensity immediately surrounding the inoculation site in the anterior right frontal lobe. (Reprinted by permission from the American Society for Microbiology)
Recent examples of applications of molecular neuroimaging in transgenic animals
Imaging of Molecular Interactions or Events
Some interesting variations on standard reporter gene assays described previously have also been adapted recently for imaging of molecular interactions in the brains of mice. In particular, imaging interacting protein partners or protein trafficking in mice could pave the way for functional proteomics in whole animals and the assessment of dysfunctional signaling networks in diseased cells and could provide a tool for evaluation of new pharmaceuticals targeted to modulate protein-protein interactions and protein translocation.56 To this end, Kim et al57 have developed a genetically encoded bioluminescent indicator for imaging the nuclear trafficking of target proteins in vivo. The principle is based on reconstitution of split fragments of Renilla luciferase (Rluc) that are inactive in their split state. The N-terminal fragment of split Rluc is intentionally localized in the nucleus, whereas the C-terminal fragment joined to a particular protein (the androgen receptor in this example) is in the cytosol. Translocation of the receptor (on binding to 5a-dihydrotestosterone [DHT]) into the nucleus results in reconstitution of full-length Rluc and recovering its bioluminescence activity. Thus, imaging and quantifying the occurrence of nucleocytoplasmic trafficking of the androgen receptor was demonstrated after brain implantation of COS-7 cells cotransfected with the genes encoding the receptor and the split Rluc fragments. On delivery of DHT, there was restored bioluminescence signal intensity indirectly indicating the trafficking of the receptor to the nucleus. This was reduced or inhibited on intraperitoneal injection of 2 agents, procymidone and polychlorinated biphenyls, supporting their likely antiandrogenic and neurotoxic effects, respectively. This study could provide a basis for a wide variety of imaging applications for screening drugs or neurotoxic compounds and testing them in preclinical animal models.
Clinical Applications in Molecular Neuroimaging Using Reporter Genes
One expectation of the ongoing developmental research in reporter gene expression imaging exemplified previously might be its straightforward translation from animal work to clinical practice.58 However, human applications present more theoretic and practical challenges than those in laboratory rodents.1 This is mostly because of the need for molecular probes to be biocompatible in humans, the presence of many physiologic/morphologic barriers to the delivery of genes and probes, and the need to develop special in vivo amplification strategies for low-level biologic events. Moreover, clinical imaging systems must be capable of obtaining high spatial/temporal resolution images and must be sensitive enough to detect these biologic processes. Because it is necessary to transduce living cells with imaging reporter genes, it follows that many of these practical requirements for successful implementation of reporter gene imaging in patients would mirror many of the logistic requirements and concerns in the field of human gene therapy. In addition, gene therapy is one of the main target areas of reporter gene imaging research because these imaging technologies are anticipated to be of significant help in monitoring transgene expression in a noninvasive manner.
Gene therapy has been one of the great yet unfulfilled promises of recent years. Yet, it has shown slow but steady progress thus far, with many of the obstacles becoming surmountable.59 Progress in this state of affairs will define mostly to what extent reporter gene expression imaging will translate into clinical practice. Overcoming the hurdles of targeting expression of exogenous or endogenous genes to cells or tissue in humans by using imaging reporters for long-term imaging is a theoretic major hurdle at present and will remain so until the current practical challenges of human gene therapy discussed previously are addressed appropriately. Intensive ongoing efforts are also underway to develop alternative simpler strategies for potential future human applications, such as the delivery of circulating exogenous split reporter proteins into cells by using leader peptide sequences. On the other hand and unlike gene therapy, future clinical applications in cell therapy (eg, by using cell-mediated immunotherapy60 or stem cells61) likely stand to benefit considerably and much sooner from reporter gene imaging, as is already clear from studies of cell trafficking in animal experiments. Indeed, Yaghoubi et al62 have recently demonstrated the first clinical experience in positron-emission tomography (PET) imaging of herpes simplex virus type-1–thymidine kinase (HSV1-tk)-expressing autologous cytolytic T lymphocytes directed at recurrent gliomas.
As with gene therapy, one of the challenges facing reporter gene expression imaging is to generate disease- or site-specific imaging strategies. Both the transductional targeting of the vector and the restriction of reporter gene expression solely to the target are potential avenues to follow once translated into clinical practice. Sufficient imaging probe would need to reach the target in vivo to achieve this specificity. Unlike MR imaging and bioluminescence imaging, both PET and SPECT use trace amounts (nonpharmacologic nanogram levels) of molecular probe to obtain images, as is the current practice for clinical scintigraphic imaging. These amounts of molecular probe are known to be safe in humans. More specifically in this regard, Yaghoubi et al63,64 have studied the 9-[4-fluorine-18 fluoro-3-(hydroxymethyl)butyl]guanine (18F- FHBG) reporter probe used for imaging expression of the HSV1-tk reporter gene. They demonstrated good kinetics, biodistribution, stability, dosimetry, and safety of 18F-FHBG in healthy human volunteers, in preparation for future imaging of patients undergoing HSV1-tk suicide gene therapy.
Unfortunately, the disadvantages of optical imaging (discussed in the previous article8) in terms of translation to clinical practice far outweigh for now its exceedingly advantageous high sensitivity for detecting low-level biologic events. Similarly, the limitations of MR imaging8 preclude for now its applications in reporter gene imaging of the brain. These issues, therefore, tend to favor the use of PET and SPECT imaging as a viable compromise for clinical implementation of this molecular imaging strategy in neuroimaging, particularly when considering the many merits of these 2 techniques.8 However, biologic and biophysical factors involved in the biodistribution of reporter probes that are potentially applicable to clinical imaging will also need to be studied carefully as they are scaled up from small animal imaging.
Regrettably, the molecular probes for the HSV1-tk enzyme (both of those based on radiolabeled uracil nucleosides and acycloguanosine derivatives) barely penetrate the intact blood-brain barrier (BBB).63,65 The BBB is a selective barrier formed by endothelial cells lining cerebral microvessels.65,66 It acts as a de facto “physical barrier” on account of complex tight junctions between adjacent endothelial cells, forcing most molecular traffic to take a transcellular route across the BBB, rather than moving paracellularly through the junctions, as in most endothelia.67 This effectively filters most ionized water-soluble molecules >180 Da in molecular weight.68 Most molecular probes for HSV1-tk are based on the structure of ganciclovir.69 This has a molecular weight of 255 Da, and it only achieves a concentration in the brain of about 50% of the plasma level. 18F-labeled acycloguanosine derivatives are heavier and usually with extra methyl and fluoro side chains added to the ganciclovir structure.
Not surprisingly, Hospers et al70 have found in previous biodistribution studies that the uptake of 9-[(3-[18F] fluoro-1-hydroxy-2-propoxy)methyl]guanine (FHPG) in brain tissue is approximately eightfold lower than the level of FHPG in plasma, reflecting this restricted passage through the intact BBB. On the other hand, the disrupted blood-tumor barrier has been shown in some studies to allow passage of similar probes in experimental rodent intracranial tumors,14 but this is not a consistent observation (see below findings in the clinical setting). As well, the permeability of the BBB may be altered during cerebral infection (eg, herpes simplex encephalitis) due to the release of chemical mediators such as bradykinin, arachidonic acid, histamine, and free radicals.71,72 Attempts to modulate the permeability of the BBB pharmacologically have been undertaken to enhance chemotherapeutic drug delivery within the brain. LeMay et al73 have previously demonstrated that the vasodilatory bradykinin analog RMP-7 increases brain tumor permeability to ganciclovir. It remains to be investigated whether the use of osmotic disruption or RMP-7 may possibly increase delivery of other HSV1-tk substrates across the BBB for molecular neuroimaging purposes.
Unfortunately, once injected systemically, the promiscuous tropism of certain viruses does limit cell-specific gene delivery by these vectors. Viral engineering strategies could ultimately benefit reporter gene expression in the clinical setting, especially to address the tropism of adenoviral vectors.74 Therefore, local delivery of imaging genes to the brain has been the only means of tissue transduction in the 2 preliminary clinical neuroimaging studies reported so far. Jacobs et al75 intraoperatively infused liposome vectors carrying the reporter HSV1-tk gene directly into tumors during a clinical phase I/II gene therapy trial of 5 patients with recurrent glioblastoma. Noninvasive primary end point (indirect) molecular imaging of the transduced “tissue dose” of vector-mediated therapeutic gene expression was performed by using the molecular probe 2′-fluoro-2′deoxy-l-beta-d-arabinofuranosyl-5-iodo-uracil (124I-FIAU) and PET. The imaged “tissue dose” of therapeutic gene expression was also correlated with the induced therapeutic effect by secondary end point molecular imaging of the metabolic activity and proliferative activity of the tumors by using [18F]fluorodeoxyglucose (18F-FDG) and 11C-methionine (11C-MET), respectively, and PET. One of the 5 patients demonstrated 124I-FIAU accumulation that was significantly above the prevector baseline and therefore consistent with successful imaging of HSV1-tk gene expression in gene therapy in man. Moreover, their findings possibly indicate that PET would be a useful tool to monitor transgene expression in gene therapy clinical trials by using viral vectors.
These early clinical examples also demonstrate the kind of synergy necessary between direct molecular imaging (eg, by using 18F-FDG and 11C-MET) and reporter gene methods in successful establishment of safe and effective gene therapy protocols in clinical practice.76 In another recent study, Dempsey et al 77 attempted, for the first time, to image expression of HSV 1716 during oncolytic viral therapy in human malignant glioma. 123I-FIAU brain SPECT imaging was undertaken in 8 patients receiving intratumoral injection of virus, but no molecular probe accumulation was detected in these treated gliomas. The authors discussed the many factors that may have contributed to this lack of imaging signal intensity, including impermeability of the BBB, inconsistent disruption of BBB, short half-life of 123I, lower sensitivity of SPECT compared with PET, the use of weak promoters, the need for more sensitive molecular probes, possible insufficient viral replication, and potential for improved administration of virus (eg, by using convection enhanced delivery). Nonetheless, this study was useful in highlighting the possible limitations of this technique and the many potential areas that need to be investigated in future research.
Future Outlook
In this article, we discussed the principles and recent technologic advances in molecular imaging of reporter gene expression in the brain. This approach is emerging as a valuable tool for monitoring the expression of genes in laboratory animals and humans. Further development of newer (eg, Gaussia luciferase78) and more sensitive and selective reporters (eg, red-shifted Rluc, with greater stability and higher light emission than native Rluc79), combined with improvements in detection technology, will consolidate the position of reporter gene imaging as a versatile method for understanding of intracellular biologic processes and the molecular basis of neurologic disorders.
Many developments in reporter gene expression imaging are anticipated during the next decade. Significant conceptual and technologic advances will most likely be seen across the 5 main general requirements for molecular imaging discussed previously and in greater detail elsewhere,1—that is, knowledge of molecular targets, availability of molecular probes, overcoming delivery barriers, developing amplification strategies, and availability of appropriate instrumentation. In particular, new strategies to circumvent the normal BBB or that target a blood-tumor barrier by the use of novel carrier vehicles (eg, in rabbits, various nutrient transporters continue to be tested across the blood-retinal barrier [used as a model of the BBB] to enhance drug bioavailability across membranes with poor permeability80) and local vasodilation or osmotic opening all merit attention, as well as the design of newer reporter gene/probe systems tailored to molecular neuroimaging. As an example of the latter, Majumdar et al81 have tested modified dipeptide monoester ganciclovir prodrugs for their greater solubility and permeability. On the other hand, Magrassi et al82 exploited the fact that HSV1-tk is not enantioselective and can therefore efficiently phosphorylate both d and l enantiomers of β-thymidine. Using autoradiography, they showed that tritiated l-β-thymidine is selectively retained to a significant extent in experimental intracranial gliomas. It has the advantage of generating less-toxic metabolites than with use of conventional HSV1-tk probes; and with appropriate radioisotopic labeling (eg, with 11C) of l-thymidine, it might be possible to adapt it for future use in PET studies of the brain.
Another study that exemplifies a search for new reporter systems with capabilities of imaging processes behind an intact BBB is that of Doubrovin et al,83 who investigated Escherichia coli xanthine phosphoribosyltransferase for nuclear imaging with radiolabeled xanthine. Again, by using autoradiography, they found that 14C xanthine was capable of specific accumulation in transfected infiltrative brain tumors. These and future similar innovations bode well for more widespread experimental and potential clinical applications of reporter gene imaging in the brain.
The merger of molecular biology and medical imaging is facilitating rapid growth of this new field by providing methods to monitor an ever-increasing number of cellular/molecular events adapted from conventional molecular assays, including reporter gene assays. Further developments will provide us with the ability to perform simultaneous imaging of multiple molecular events in 1 population of cells in living subjects. This may be attainable by combining 2 or more of the previously described strategies for gene marking and imaging the trafficking of cells with those entailing linked expression of an imaging gene to an endogenous promoter or to an exogenous therapeutic gene. As such, in these applications, it is foreseeable that 1 reporter may reveal the spatial distribution of cells and whether they have reached a specific target, and another reporter may indicate whether a certain gene becomes upregulated at this site or if a more complex interaction occurs. These endeavors will be aided by the availability of multiple fusion reporter constructs (eg, those that combine PET/bioluminescence/fluorescence imaging capabilities in 1 gene),84 the use of which should accelerate the validation of reporter gene approaches developed in cell culture for translation into preclinical models and subsequent clinical imaging of neurologic disorders. With continued rapid advancements in this field, the experimental and clinical neurosciences stand to gain considerably from noninvasive molecular imaging of the expression of multiple fused reporter genes by using multiple imaging techniques.85 These approaches are likely to play an increasingly important role in defining molecular events in the field of cancer biology, cell biology, and gene therapy within the central nervous system.
References
- Copyright © American Society of Neuroradiology